Nef induces multiple genes involved in cholesterol synthesis and uptake in human immunodeficiency virus type 1-infected T cells
- PMID: 16014965
- PMCID: PMC1181597
- DOI: 10.1128/JVI.79.15.10053-10058.2005
Nef induces multiple genes involved in cholesterol synthesis and uptake in human immunodeficiency virus type 1-infected T cells
Abstract
Several recent reports indicate that cholesterol might play an important role in human immunodeficiency virus type 1 (HIV-1) replication. We investigated the effects of HIV-1 infection on cholesterol biosynthesis and uptake using microarrays. HIV-1 increased gene expression of cholesterol genes in both transformed T-cell lines and primary CD4(+) T cells. Consistent with our microarray data, (14)C-labeled mevalonate and acetate incorporation was increased in HIV-1-infected cells. Our data also demonstrate that changes in cholesterol biosynthesis and uptake are only observed in the presence of functional Nef, suggesting that increased cholesterol synthesis may contribute to Nef-mediated enhancement of virion infectivity and viral replication.
Figures


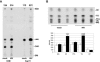
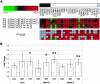
References
-
- Folch, J., M. Lees, and G. H. Sloane Stanley. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226:497-509. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

