Variants of CCR5, which are permissive for HIV-1 infection, show distinct functional responses to CCL3, CCL4 and CCL5
- PMID: 16015368
- PMCID: PMC1369982
- DOI: 10.1038/sj.gene.6364247
Variants of CCR5, which are permissive for HIV-1 infection, show distinct functional responses to CCL3, CCL4 and CCL5
Abstract
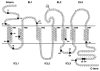
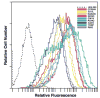
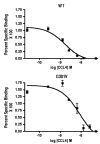
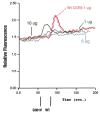
CCR5 is one of the primary coreceptors for Env-mediated fusion between cells and human immunodeficiency virus type 1 (HIV-1). Analyses of CCR5 variants in cohorts of HIV-1 high-risk individuals led to the identification of multiple single amino-acid substitutions, which may have functional consequences. This study focused on eight naturally occurring allelic variants located between amino-acid residues 60 and 334 of CCR5. All studied allelic variants were highly expressed on the cell surface of HEK-293 cells and permissive for HIV-1 infection. Variant G301V showed 3.5-fold increase in 50% effective concentration (EC(50)) for CCL4 (MIP 1beta) in a competitive binding assay. There was also a significant reduction in CCL5 (RANTES) EC(50) for the R223Q, A335V and Y339F variants. The most unexpected functional abnormality was exhibited by the R60S variant that exhibited a loss of ligand-induced desensitization in chemotaxis assays, but showed normal CCL4 and CCL5 binding avidity. This mutation is located in the first intracellular loop, a domain that has not previously been shown to be involved in receptor desensitization. In conclusion, our results support earlier studies showing that these naturally occurring point mutations do not limit HIV-1 infection, and indicated that single amino-acid changes can have unexpected functional consequences.
Genes and Immunity (2005) 6, 609-619. doi:10.1038/sj.gene.6364247; published online 14 July 2005.
Figures


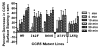

Similar articles
-
Naturally occurring CCR5 extracellular and transmembrane domain variants affect HIV-1 Co-receptor and ligand binding function.J Biol Chem. 1999 Jun 4;274(23):16228-34. doi: 10.1074/jbc.274.23.16228. J Biol Chem. 1999. PMID: 10347178
-
Aminooxypentane addition to the chemokine macrophage inflammatory protein-1alpha P increases receptor affinities and HIV inhibition.J Biol Chem. 2000 Dec 15;275(50):39254-61. doi: 10.1074/jbc.M006768200. J Biol Chem. 2000. PMID: 11005816
-
Involvement of both the V2 and V3 regions of the CCR5-tropic human immunodeficiency virus type 1 envelope in reduced sensitivity to macrophage inflammatory protein 1alpha.J Virol. 2000 Feb;74(4):1787-93. doi: 10.1128/jvi.74.4.1787-1793.2000. J Virol. 2000. PMID: 10644351 Free PMC article.
-
Molecular anatomy of CCR5 engagement by physiologic and viral chemokines and HIV-1 envelope glycoproteins: differences in primary structural requirements for RANTES, MIP-1 alpha, and vMIP-II Binding.J Mol Biol. 2001 Nov 9;313(5):1181-93. doi: 10.1006/jmbi.2001.5086. J Mol Biol. 2001. PMID: 11700073
-
The role of CCR5 chemokine ligands and antibodies to CCR5 coreceptors in preventing HIV infection.Trends Immunol. 2002 Jul;23(7):347-51. doi: 10.1016/s1471-4906(02)02252-4. Trends Immunol. 2002. PMID: 12103354 Review. No abstract available.
Cited by
-
Immune-Based Prediction of COVID-19 Severity and Chronicity Decoded Using Machine Learning.Front Immunol. 2021 Jun 28;12:700782. doi: 10.3389/fimmu.2021.700782. eCollection 2021. Front Immunol. 2021. PMID: 34262570 Free PMC article.
-
Pro-inflammatory macrophages suppress HIV replication in humanized mice and ex vivo co-cultures.Front Immunol. 2024 Nov 7;15:1439328. doi: 10.3389/fimmu.2024.1439328. eCollection 2024. Front Immunol. 2024. PMID: 39575258 Free PMC article.
-
mRNA transfection of a novel TAL effector nuclease (TALEN) facilitates efficient knockout of HIV co-receptor CCR5.Nucleic Acids Res. 2015 Jun 23;43(11):5560-71. doi: 10.1093/nar/gkv469. Epub 2015 May 11. Nucleic Acids Res. 2015. PMID: 25964300 Free PMC article.
-
Mapping Interaction Sites on Human Chemokine Receptors by Deep Mutational Scanning.J Immunol. 2018 Jun 1;200(11):3825-3839. doi: 10.4049/jimmunol.1800343. Epub 2018 Apr 20. J Immunol. 2018. PMID: 29678950 Free PMC article.
-
CCL4 enhances preosteoclast migration and its receptor CCR5 downregulation by RANKL promotes osteoclastogenesis.Cell Death Dis. 2018 May 1;9(5):495. doi: 10.1038/s41419-018-0562-5. Cell Death Dis. 2018. PMID: 29717113 Free PMC article.
References
-
- Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor [see comments] Science. 1996;272:872–877. - PubMed
-
- Moore JP, Trkola A, Dragic T. Co-receptors for HIV-1 entry. Curr Opin Immunol. 1997;9:551–562. - PubMed
-
- Littman DR. Chemokine receptors: keys to AIDS pathogenesis? Cell. 1998;93:677–680. - PubMed
-
- Raport CJ, Gosling J, Schweickart VL, Gray PW, Charo IF. Molecular cloning and functional characterization of a novel human CC chemokine receptor (CCR5) for RANTES, MIP-1beta, and MIP-1alpha. J Biol Chem. 1996;271:17161–17166. - PubMed
-
- Zlotnik A, Yoshie O. Chemokines: a new classification system and their role in immunity. Immunity. 2000;12:121–127. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

