Thermal aggregation of SARS-CoV membrane protein
- PMID: 16023741
- PMCID: PMC7112854
- DOI: 10.1016/j.jviromet.2005.05.022
Thermal aggregation of SARS-CoV membrane protein
Abstract
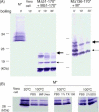
SARS-CoV membrane protein could be detected easily using Western blotting in non-denaturing condition but not regular denaturing treatment. Boiling treatment, causing the aggregation of SARS-CoV membrane protein in the stacking gels, results in the failure to detect the membrane protein in the separating gels. Aggregated membrane proteins could not be dissociated by 1% Triton-X 100, 6M urea, or 2% SDS. The region with amino acid residues from 51 to 170 is responsible for thermal aggregation of SARS-CoV membrane protein. Hydrophobic regions with amino acid residues from 61 to 90, from 91 to 100, from 136 to 170, are essential for this protein aggregation. Thermal aggregation of SARS-CoV membrane protein is not unique among structural proteins of coronaviruses. However, SARS-CoV membrane protein seems to be more sensitive to heat treatment, since the membrane protein of MHV-JHM, another member of the Coronaviridae, would not aggregate after the same treatment. Therefore, if SARS-CoV membrane protein needs to be analyzed using SDS-PAGE, boiling should be avoided. Thermal aggregation of SARS-CoV membrane protein may be one of the reasons for the inactivation of this virus by heat. The unusual property of SARS-CoV membrane protein aggregation induced by heat also provides a model for the study of protein aggregation.
Figures


Similar articles
-
Studies on membrane topology, N-glycosylation and functionality of SARS-CoV membrane protein.Virol J. 2009 Jun 18;6:79. doi: 10.1186/1743-422X-6-79. Virol J. 2009. PMID: 19534833 Free PMC article.
-
Interactions between M protein and other structural proteins of severe, acute respiratory syndrome-associated coronavirus.J Biomed Sci. 2008 Nov;15(6):707-17. doi: 10.1007/s11373-008-9278-3. Epub 2008 Sep 16. J Biomed Sci. 2008. PMID: 18792806 Free PMC article.
-
The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles.J Virol. 2008 Nov;82(22):11318-30. doi: 10.1128/JVI.01052-08. Epub 2008 Aug 27. J Virol. 2008. PMID: 18753196 Free PMC article.
-
Properties of Coronavirus and SARS-CoV-2.Malays J Pathol. 2020 Apr;42(1):3-11. Malays J Pathol. 2020. PMID: 32342926 Review.
-
An overall picture of SARS coronavirus (SARS-CoV) genome-encoded major proteins: structures, functions and drug development.Curr Pharm Des. 2006;12(35):4539-53. doi: 10.2174/138161206779010459. Curr Pharm Des. 2006. PMID: 17168760 Review.
Cited by
-
Modulation of the unfolded protein response by the severe acute respiratory syndrome coronavirus spike protein.J Virol. 2006 Sep;80(18):9279-87. doi: 10.1128/JVI.00659-06. J Virol. 2006. PMID: 16940539 Free PMC article.
-
Small interfering RNA effectively inhibits the expression of SARS coronavirus membrane gene at two novel targeting sites.Molecules. 2010 Oct 18;15(10):7197-207. doi: 10.3390/molecules15107197. Molecules. 2010. PMID: 20956884 Free PMC article.
-
Thermal inactivation scaling applied for SARS-CoV-2.Biophys J. 2021 Mar 16;120(6):1054-1059. doi: 10.1016/j.bpj.2020.11.2259. Epub 2020 Nov 28. Biophys J. 2021. PMID: 33253633 Free PMC article.
-
Physicochemical properties of SARS-CoV-2 for drug targeting, virus inactivation and attenuation, vaccine formulation and quality control.Electrophoresis. 2020 Jul;41(13-14):1137-1151. doi: 10.1002/elps.202000121. Epub 2020 Jun 8. Electrophoresis. 2020. PMID: 32469436 Free PMC article. Review.
-
Analytical modeling of three-stage inactivation of viruses within droplets and solid porous particles.Eur Phys J Plus. 2021;136(6):663. doi: 10.1140/epjp/s13360-021-01651-1. Epub 2021 Jun 17. Eur Phys J Plus. 2021. PMID: 34155467 Free PMC article.
References
-
- Callebaut P.E., Pensaert M.B. Characterization and isolation of structural polypeptides in haemagglutinating encephalomyelitis virus. J. Gen. Virol. 1980;48(1):193–204. - PubMed
-
- de Haan C.A., Roestenberg P., de Wit M., de Vries A.A., Nilsson T., Vennema H., Rottier P.J. Structural requirements for O-glycosylation of the mouse hepatitis virus membrane protein. J. Biol. Chem. 1998;273(45):29905–29914. - PubMed
-
- Deregt D., Sabara M., Babiuk L.A. Structural proteins of bovine coronavirus and their intracellular processing. J. Gen. Virol. 1987;68(Pt 11):2863–2877. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

