A Rik1-associated, cullin-dependent E3 ubiquitin ligase is essential for heterochromatin formation
- PMID: 16024659
- PMCID: PMC1176008
- DOI: 10.1101/gad.1328005
A Rik1-associated, cullin-dependent E3 ubiquitin ligase is essential for heterochromatin formation
Abstract
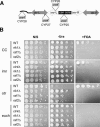
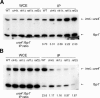
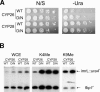
Heterochromatin is critical for proper centromere and telomere function, and it plays a key role in the transcriptional silencing of specific genomic loci. In fission yeast, the Rik1 protein functions with the Clr4 histone methyltransferase at an early step in heterochromatin formation. Here, we use mass spectrometry and tandem affinity purification of a Rik1-TAP fusion protein to identify Rik1-associated proteins. These studies identify two novel proteins, Raf1 and Raf2, which we find are required for H3-K9 methylation and for transcriptional silencing within centromeric heterochromatin. We also find that subunits of a cullin-dependent E3 ubiquitin ligase are associated with Rik1 and Clr4, and Rik1-TAP preparations exhibit robust E3 ubiquitin ligase activity. Furthermore, expression of a dominant-negative allele of the Pcu4 cullin subunit disrupts regulation of K4 methylation within heterochromatin. These studies provide evidence for a novel Rik1-associated E3 ubiquitin ligase that is required for heterochromatin formation.
Figures



References
-
- Allshire R.C., Javerzat, J.P., Redhead, N.J., and Cranston, G. 1994. Position effect variegation at fission yeast centromeres. Cell 76: 157–169. - PubMed
-
- Allshire R.C., Nimmo, E.R., Ekwall, K., Javerzat, J.P., and Cranston, G. 1995. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes & Dev. 9: 218–233. - PubMed
-
- Bahler J., Wu, J.Q., Longtine, M.S., Shah, N.G., McKenzie III, A., Steever, A.B., Wach, A., Philippsen, P., and Pringle, J.R. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951. - PubMed
-
- Bernard P., Maure, J.F., Partridge, J.F., Genier, S., Javerzat, J.P., and Allshire, R.C. 2001. Requirement of heterochromatin for cohesion at centromeres. Science 294: 2539–2542. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
