A-tract clusters may facilitate DNA packaging in bacterial nucleoid
- PMID: 16024741
- PMCID: PMC1176013
- DOI: 10.1093/nar/gki699
A-tract clusters may facilitate DNA packaging in bacterial nucleoid
Abstract
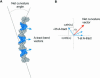
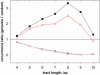
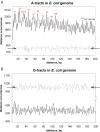
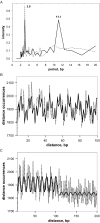
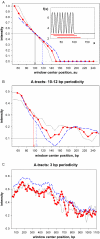
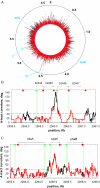
Molecular mechanisms of bacterial chromosome packaging are still unclear, as bacteria lack nucleosomes or other apparent basic elements of DNA compaction. Among the factors facilitating DNA condensation may be a propensity of the DNA molecule for folding due to its intrinsic curvature. As suggested previously, the sequence correlations in genome reflect such a propensity [Trifonov and Sussman (1980) Proc. Natl Acad. Sci. USA, 77, 3816-3820]. To further elaborate this concept, we analyzed positioning of A-tracts (the sequence motifs introducing the most pronounced DNA curvature) in the Escherichia coli genome. First, we observed that the A-tracts are over-represented and distributed 'quasi-regularly' throughout the genome, including both the coding and intergenic sequences. Second, there is a 10-12 bp periodicity in the A-tract positioning indicating that the A-tracts are phased with respect to the DNA helical repeat. Third, the phased A-tracts are organized in approximately 100 bp long clusters. The latter feature was revealed with the help of a novel approach based on the Fourier series expansion of the A-tract distance autocorrelation function. Since the A-tracts introduce local bends of the DNA duplex and these bends accumulate when properly phased, the observed clusters would facilitate DNA looping. Also, such clusters may serve as binding sites for the nucleoid-associated proteins that have affinities for curved DNA (such as HU, H-NS, Hfq and CbpA). Therefore, we suggest that the approximately 100 bp long clusters of the phased A-tracts constitute the 'structural code' for DNA compaction by providing the long-range intrinsic curvature and increasing stability of the DNA complexes with architectural proteins.
Figures


Similar articles
-
The architectural role of nucleoid-associated proteins in the organization of bacterial chromatin: a molecular perspective.J Struct Biol. 2006 Nov;156(2):262-72. doi: 10.1016/j.jsb.2006.05.006. Epub 2006 Jun 3. J Struct Biol. 2006. PMID: 16879983 Review.
-
Investigating the interplay between nucleoid-associated proteins, DNA curvature, and CRISPR elements using comparative genomics.PLoS One. 2014 Mar 3;9(3):e90940. doi: 10.1371/journal.pone.0090940. eCollection 2014. PLoS One. 2014. PMID: 24595272 Free PMC article.
-
A DNA structural atlas for Escherichia coli.J Mol Biol. 2000 Jun 16;299(4):907-30. doi: 10.1006/jmbi.2000.3787. J Mol Biol. 2000. PMID: 10843847
-
Archaeal histone selection of nucleosome positioning sequences and the procaryotic origin of histone-dependent genome evolution.J Mol Biol. 2000 Oct 13;303(1):25-34. doi: 10.1006/jmbi.2000.4128. J Mol Biol. 2000. PMID: 11021967
-
[The bacterial nucleoid].Rev Latinoam Microbiol. 1995 Jul-Sep;37(3):281-90. Rev Latinoam Microbiol. 1995. PMID: 8850347 Review. Spanish.
Cited by
-
Bacterial nucleoid is a riddle wrapped in a mystery inside an enigma.J Bacteriol. 2024 Mar 21;206(3):e0021123. doi: 10.1128/jb.00211-23. Epub 2024 Feb 15. J Bacteriol. 2024. PMID: 38358278 Free PMC article. Review.
-
Involvement of the leucine response transcription factor LeuO in regulation of the genes for sulfa drug efflux.J Bacteriol. 2009 Jul;191(14):4562-71. doi: 10.1128/JB.00108-09. Epub 2009 May 8. J Bacteriol. 2009. PMID: 19429622 Free PMC article.
-
Assessing diversity of DNA structure-related sequence features in prokaryotic genomes.DNA Res. 2014 Jun;21(3):285-97. doi: 10.1093/dnares/dst057. Epub 2014 Jan 9. DNA Res. 2014. PMID: 24408877 Free PMC article.
-
Genome-wide analysis of Fis binding in Escherichia coli indicates a causative role for A-/AT-tracts.Genome Res. 2008 Jun;18(6):900-10. doi: 10.1101/gr.070276.107. Epub 2008 Mar 13. Genome Res. 2008. PMID: 18340041 Free PMC article.
-
Amyloid-like Hfq interaction with single-stranded DNA: involvement in recombination and replication in Escherichia coli.QRB Discov. 2022 Sep 7;3:e15. doi: 10.1017/qrd.2022.15. eCollection 2022. QRB Discov. 2022. PMID: 37529279 Free PMC article.
References
-
- Pettijohn D.E., Sinden R.R. Structure of the isolated nucleoid. In: Nanninga N., editor. Molecular Cytology of Escherichia coli. London: Academic Press; 1985. pp. 199–227.
-
- Case R.B., Chang Y.P., Smith S.B., Gore J., Cozzarelli N.R., Bustamante C. The bacterial condensin MukBEF compacts DNA into a repetitive, stable structure. Science. 2004;305:222–227. - PubMed
-
- Kleppe K., Ovrebo S., Lossius I. The bacterial nucleoid. J. Gen. Microbiol. 1979;112:1–13. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

