DNA replication origin plasticity and perturbed fork progression in human inverted repeats
- PMID: 16024811
- PMCID: PMC1190330
- DOI: 10.1128/MCB.25.15.6789-6797.2005
DNA replication origin plasticity and perturbed fork progression in human inverted repeats
Abstract
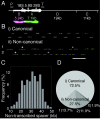
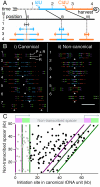
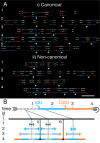
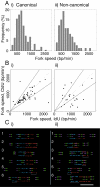
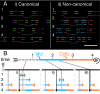
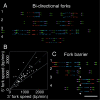
The stability of metazoan genomes during their duplication depends on the spatiotemporal activation of origins and the progression of forks. Human rRNA genes represent a unique challenge to DNA replication since a large proportion of them exist as noncanonical palindromes in addition to canonical tandem repeats. Whether origin usage and/or fork elongation can cope with the variable structure of these genes is unknown. By analyzing single combed DNA molecules from HeLa cells, we studied the rRNA gene replication program according to the organization of canonical versus noncanonical rRNA genes. Origin positioning, spacing, and timing were not affected by the underlying rRNA gene physical structure. Conversely, fork arrest, both temporary and permanent, occurred more frequently when rRNA gene palindromes were encountered. These findings reveal that while initiation mechanisms are flexible enough to adapt to an rRNA gene structure of any arrangement, palindromes represent obstacles to fork progression, which is a likely source of genomic instability.
Figures
Similar articles
-
Impairment of replication fork progression mediates RNA polII transcription-associated recombination.EMBO J. 2005 Mar 23;24(6):1267-76. doi: 10.1038/sj.emboj.7600602. Epub 2005 Mar 3. EMBO J. 2005. PMID: 15775982 Free PMC article.
-
Unscheduled DNA replication origin activation at inserted HPV 18 sequences in a HPV-18/MYC amplicon.Genes Chromosomes Cancer. 2007 Aug;46(8):724-34. doi: 10.1002/gcc.20448. Genes Chromosomes Cancer. 2007. PMID: 17444495
-
Human ribosomal RNA gene arrays display a broad range of palindromic structures.Genome Res. 2005 Aug;15(8):1079-85. doi: 10.1101/gr.3970105. Epub 2005 Jul 15. Genome Res. 2005. PMID: 16024823 Free PMC article.
-
Stalled replication forks: making ends meet for recognition and stabilization.Bioessays. 2010 Aug;32(8):687-97. doi: 10.1002/bies.200900196. Bioessays. 2010. PMID: 20658707 Review.
-
[G-quadruplex: key controllers of human genome duplication].Med Sci (Paris). 2017 Dec;33(12):1063-1070. doi: 10.1051/medsci/20173312013. Epub 2017 Dec 20. Med Sci (Paris). 2017. PMID: 29261494 Review. French.
Cited by
-
The Human RNA Polymerase I Transcription Terminator Complex Acts as a Replication Fork Barrier That Coordinates the Progress of Replication with rRNA Transcription Activity.Mol Cell Biol. 2015 May;35(10):1871-81. doi: 10.1128/MCB.01521-14. Epub 2015 Mar 16. Mol Cell Biol. 2015. PMID: 25776556 Free PMC article.
-
Segmental duplications and evolutionary plasticity at tumor chromosome break-prone regions.Genome Res. 2008 Mar;18(3):370-9. doi: 10.1101/gr.7010208. Epub 2008 Jan 29. Genome Res. 2008. PMID: 18230801 Free PMC article.
-
Molecular Combing of Single DNA Molecules on the 10 Megabase Scale.Sci Rep. 2016 Jan 19;6:19636. doi: 10.1038/srep19636. Sci Rep. 2016. PMID: 26781994 Free PMC article.
-
A Disease-Causing Single Amino Acid Deletion in the Coiled-Coil Domain of RAD50 Impairs MRE11 Complex Functions in Yeast and Humans.Cell Rep. 2020 Dec 29;33(13):108559. doi: 10.1016/j.celrep.2020.108559. Cell Rep. 2020. PMID: 33378670 Free PMC article.
-
Variability of Human rDNA.Cells. 2021 Jan 20;10(2):196. doi: 10.3390/cells10020196. Cells. 2021. PMID: 33498263 Free PMC article. Review.
References
-
- Anglana, M., F. Apiou, A. Bensimon, and M. Debatisse. 2003. Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell 114:385-394. - PubMed
-
- Bensimon, A., A. Simon, A. Chiffaudel, V. Croquette, F. Heslot, and D. Bensimon. 1994. Alignment and sensitive detection of DNA by a moving interface. Science 265:2096-2098. - PubMed
-
- Brewer, B. J., and W. L. Fangman. 1988. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell 55:637-643. - PubMed
-
- Brewer, B. J., D. Lockshon, and W. L. Fangman. 1992. The arrest of replication forks in the rDNA of yeast occurs independently of transcription. Cell 71:267-276. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
