The scale of mutational variation in the murid genome
- PMID: 16024822
- PMCID: PMC1182221
- DOI: 10.1101/gr.3895005
The scale of mutational variation in the murid genome
Abstract
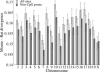
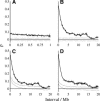
Mutation rates vary across mammalian genomes, but little is known about the scale over which this variation occurs. Knowledge of the magnitude and scale of mutational variation is required to understand the processes that drive mutation, and is essential in formulating a robust null hypothesis for comparative genomics studies. Here we estimate the scale of mutational variation in the murid genome by calculating the spatial autocorrelation of nucleotide substitution rates in ancestral repeats. Such transposable elements are good candidates for neutrally evolving sequence and therefore well suited for the study of mutation rate variation. We find that the autocorrelation coefficient decays to a value close to zero by approximately 15 Mb, with little apparent variation in mutation rate under 100 kb. We conclude that the primary scale over which mutation rates vary is subchromosomal. Furthermore, our analysis shows that within-chromosome mutational variability exceeds variation among chromosomes by approximately one order of magnitude. Thus, differences in mutation rate between different regions of the same chromosome frequently exceed differences both between whole autosomes and between autosomes and the X-chromosome. Our results indicate that factors other than the time spent in the male germ line are important in driving mutation rates. This raises questions about the biological mechanism(s) that produce new mutations and has implications for the study of male-driven evolution.
Figures




References
-
- Arndt, P.F., Burge, C.B., and Hwa, T. 2003. DNA sequence evolution with neighbor-dependent mutation. J. Comput. Biol. 10: 313–322. - PubMed
-
- Bejerano, G., Pheasant, M., Makunin, I., Stephen, S., Kent, W.J., Mattick, J.S., and Haussler, D. 2004. Ultraconserved elements in the human genome. Science 304: 1321–1325. - PubMed
-
- Box, G.E.P., Jenkins, G.M., and Reinsel, G.C. 1994. Time series analysis: Forecasting and control, 3rd ed. Prentice-Hall, Upper Saddle River, NJ.
Web site references
-
- http://www.repeatmasker.org/; the program RepeatMasker is available for download from this site.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous
