The structure of the TRAPP subunit TPC6 suggests a model for a TRAPP subcomplex
- PMID: 16025134
- PMCID: PMC1369139
- DOI: 10.1038/sj.embor.7400463
The structure of the TRAPP subunit TPC6 suggests a model for a TRAPP subcomplex
Abstract
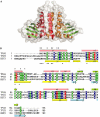
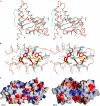
The TRAPP (transport protein particle) complexes are tethering complexes that have an important role at the different steps of vesicle transport. Recently, the crystal structures of the TRAPP subunits SEDL and BET3 have been determined, and we present here the 1.7 Angstroms crystal structure of human TPC6, a third TRAPP subunit. The protein adopts an alpha/beta-plait topology and forms a dimer. In spite of low sequence similarity, the structure of TPC6 strikingly resembles that of BET3. The similarity is especially prominent at the dimerization interfaces of the proteins. This suggests heterodimerization of TPC6 and BET3, which is shown by in vitro and in vivo association studies. Together with TPC5, another TRAPP subunit, TPC6 and BET3 are supposed to constitute a family of paralogous proteins with closely similar three-dimensional structures but little sequence similarity among its members.
Figures




Similar articles
-
Structure of the Bet3-Tpc6B core of TRAPP: two Tpc6 paralogs form trimeric complexes with Bet3 and Mum2.J Mol Biol. 2006 Aug 4;361(1):22-32. doi: 10.1016/j.jmb.2006.06.012. Epub 2006 Jun 21. J Mol Biol. 2006. PMID: 16828797
-
Structure of palmitoylated BET3: insights into TRAPP complex assembly and membrane localization.EMBO J. 2005 Mar 9;24(5):875-84. doi: 10.1038/sj.emboj.7600565. Epub 2005 Feb 3. EMBO J. 2005. PMID: 15692564 Free PMC article.
-
Crystal structure of bet3 reveals a novel mechanism for Golgi localization of tethering factor TRAPP.Nat Struct Mol Biol. 2005 Jan;12(1):38-45. doi: 10.1038/nsmb871. Epub 2004 Dec 19. Nat Struct Mol Biol. 2005. PMID: 15608655
-
The TRAPP complex: insights into its architecture and function.Traffic. 2008 Dec;9(12):2032-42. doi: 10.1111/j.1600-0854.2008.00833.x. Epub 2008 Oct 14. Traffic. 2008. PMID: 18801063 Free PMC article. Review.
-
In sickness and in health: the role of TRAPP and associated proteins in disease.Traffic. 2014 Aug;15(8):803-18. doi: 10.1111/tra.12183. Epub 2014 Jul 1. Traffic. 2014. PMID: 24917561 Review.
Cited by
-
The TRAPP complexes: discriminating GTPases in context.FEBS Lett. 2023 Mar;597(6):721-733. doi: 10.1002/1873-3468.14557. Epub 2022 Dec 21. FEBS Lett. 2023. PMID: 36481981 Free PMC article. Review.
-
An effective system for detecting protein-protein interaction based on in vivo cleavage by PPV NIa protease.Protein Cell. 2012 Dec;3(12):921-8. doi: 10.1007/s13238-012-2101-y. Epub 2012 Oct 24. Protein Cell. 2012. PMID: 23096592 Free PMC article.
-
Structural and functional analysis of the globular head domain of p115 provides insight into membrane tethering.J Mol Biol. 2009 Aug 7;391(1):26-41. doi: 10.1016/j.jmb.2009.04.062. Epub 2009 May 4. J Mol Biol. 2009. PMID: 19414022 Free PMC article.
-
TRAPP Complexes in Secretion and Autophagy.Front Cell Dev Biol. 2016 Mar 30;4:20. doi: 10.3389/fcell.2016.00020. eCollection 2016. Front Cell Dev Biol. 2016. PMID: 27066478 Free PMC article. Review.
-
Vectors for co-expression of an unrestricted number of proteins.Nucleic Acids Res. 2007;35(6):e43. doi: 10.1093/nar/gkm067. Epub 2007 Feb 20. Nucleic Acids Res. 2007. PMID: 17311810 Free PMC article.
References
-
- Budisa N, Steipe B, Demange P, Eckerskorn C, Kellermann J, Huber R (1995) High-level biosynthetic substitution of methionine in proteins by its analogs 2-aminohexanoic acid, selenomethionine, telluromethionine and ethionine in Escherichia coli. Eur J Biochem 230: 788–796 - PubMed
-
- DeLano WL (2003) The PyMOL Molecular Graphics System. DeLano Scientific LLC, San Carlos, CA, USA
-
- Gavin AC et al. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147 - PubMed
-
- Higgins DG, Bleasby AJ, Fuchs R (1992) CLUSTAL V: improved software for multiple sequence alignment. Comput Appl Biosci 8: 189–191 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

