ATRIP oligomerization is required for ATR-dependent checkpoint signaling
- PMID: 16027118
- PMCID: PMC1360181
- DOI: 10.1074/jbc.M504961200
ATRIP oligomerization is required for ATR-dependent checkpoint signaling
Abstract
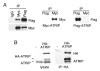
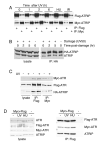
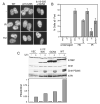
The ATM and ATR kinases signal cell cycle checkpoint responses to DNA damage. Inactive ATM is an oligomer that is disrupted to form active monomers in response to ionizing radiation. We examined whether ATR is activated by a similar mechanism. We found that the ATRIP subunit of the ATR kinase and ATR itself exist as homooligomers in cells. We did not detect regulation of ATR or ATRIP oligomerization after DNA damage. The predicted coiled-coil domain of ATRIP is essential for ATRIP oligomerization, stable ATR binding, and accumulation of ATRIP at DNA lesions. Additionally, the ATRIP coiled-coil is also required for ATRIP to support ATR-dependent checkpoint signaling to Chk1. Replacing the ATRIP coiled-coil domain with a heterologous dimerization domain restored stable binding to ATR and localization to damage-induced intranuclear foci. Thus, the ATR-ATRIP complex exists in higher order oligomeric states within cells and ATRIP oligomerization is essential for its function.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

