Mutually dependent secretion of proteins required for mycobacterial virulence
- PMID: 16030141
- PMCID: PMC1176248
- DOI: 10.1073/pnas.0504922102
Mutually dependent secretion of proteins required for mycobacterial virulence
Abstract
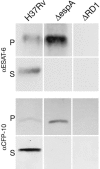
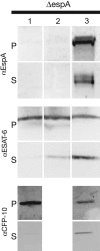
The ESX-1 locus is a region critical for full virulence in Mycobacterium tuberculosis, which encodes two secreted proteins as well as other genes involved in their secretion. The mechanism of secretion of the two proteins, ESAT-6 and CFP-10, and their function remain unknown. Using proteomic methods to search for additional proteins secreted by the ESX-1 locus, we discovered that a protein encoded by a chromosomally unlinked gene, espA, is also secreted by strains that contain the ESX-1 locus but not by strains with ESX-1 deletions. Mutations in individual ESX-1 genes, including those that encode ESAT-6 and CFP-10, were found to block EspA secretion. Surprisingly, mutants that lack espA reciprocally failed to secrete ESAT-6 and CFP-10 and were as attenuated as ESX-1 mutants in virulence assays. The results indicate that secretion of these proteins, which are each critical for virulence of pathogenic mycobacteria, is mutually dependent. The results further suggest that discerning the nature of the interaction and the structure of macromolecular complexes will provide insights into both an alternative mechanism of protein secretion and mycobacterial virulence.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
- K08 AI050734/AI/NIAID NIH HHS/United States
- HL64550/HL/NHLBI NIH HHS/United States
- AI23545/AI/NIAID NIH HHS/United States
- AI50734/AI/NIAID NIH HHS/United States
- R01 AI048704/AI/NIAID NIH HHS/United States
- HL68533/HL/NHLBI NIH HHS/United States
- AI51929/AI/NIAID NIH HHS/United States
- R01 HL064550/HL/NHLBI NIH HHS/United States
- R01 AI051929/AI/NIAID NIH HHS/United States
- R01 AI023545/AI/NIAID NIH HHS/United States
- R01 AI007118/AI/NIAID NIH HHS/United States
- R01 HL068533/HL/NHLBI NIH HHS/United States
- AI48704/AI/NIAID NIH HHS/United States
- AI07118/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

