Novel intracellular 3-hydroxybutyrate-oligomer hydrolase in Wautersia eutropha H16
- PMID: 16030206
- PMCID: PMC1196019
- DOI: 10.1128/JB.187.15.5129-5135.2005
Novel intracellular 3-hydroxybutyrate-oligomer hydrolase in Wautersia eutropha H16
Abstract
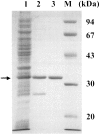
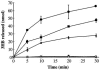
Wautersia eutropha H16 (formerly Ralstonia eutropha) mobilizes intracellularly accumulated poly(3-hydroxybutyrate) (PHB) with intracellular poly(3-hydroxybutyrate) depolymerases. In this study, a novel intracellular 3-hydroxybutyrate-oligomer hydrolase (PhaZc) gene was cloned and overexpressed in Escherichia coli. Then PhaZc was purified and characterized. Immunoblot analysis with polyclonal antiserum against PhaZc revealed that most PhaZc is present in the cytosolic fraction and a small amount is present in the poly(3-hydroxybutyrate) inclusion bodies of W. eutropha. PhaZc degraded various 3-hydroxybutyrate oligomers at a high specific activity and artificial amorphous poly(3-hydroxybutyrate) at a lower specific activity. Native PHB granules and semicrystalline PHB were not degraded by PhaZc. A PhaZ deletion mutation enhanced the deposition of PHB in the logarithmic phase in nutrient-rich medium. PhaZc differs from the hydrolases of W. eutropha previously reported and is a novel type of intracellular 3-hydroxybutyrate-oligomer hydrolase, and it participates in the mobilization of PHB along with other hydrolases.
Figures



Similar articles
-
Purification and properties of an intracellular 3-hydroxybutyrate-oligomer hydrolase (PhaZ2) in Ralstonia eutropha H16 and its identification as a novel intracellular poly(3-hydroxybutyrate) depolymerase.J Bacteriol. 2003 Jun;185(12):3485-90. doi: 10.1128/JB.185.12.3485-3490.2003. J Bacteriol. 2003. PMID: 12775684 Free PMC article.
-
Properties of a novel intracellular poly(3-hydroxybutyrate) depolymerase with high specific activity (PhaZd) in Wautersia eutropha H16.J Bacteriol. 2005 Oct;187(20):6982-90. doi: 10.1128/JB.187.20.6982-6990.2005. J Bacteriol. 2005. PMID: 16199568 Free PMC article.
-
The "intracellular" poly(3-hydroxybutyrate) (PHB) depolymerase of Rhodospirillum rubrum is a periplasm-located protein with specificity for native PHB and with structural similarity to extracellular PHB depolymerases.J Bacteriol. 2004 Nov;186(21):7243-53. doi: 10.1128/JB.186.21.7243-7253.2004. J Bacteriol. 2004. PMID: 15489436 Free PMC article.
-
Intracellular degradation of poly(3-hydroxybutyrate) granules of Zoogloea ramigera I-16-M.FEMS Microbiol Rev. 1992 Dec;9(2-4):333-8. doi: 10.1016/0378-1097(92)90327-k. FEMS Microbiol Rev. 1992. PMID: 1476778 Review.
-
New insights in the formation of polyhydroxyalkanoate granules (carbonosomes) and novel functions of poly(3-hydroxybutyrate).Environ Microbiol. 2014 Aug;16(8):2357-73. doi: 10.1111/1462-2920.12356. Epub 2014 Jan 21. Environ Microbiol. 2014. PMID: 24329995 Review.
Cited by
-
Comparative proteome analysis reveals four novel polyhydroxybutyrate (PHB) granule-associated proteins in Ralstonia eutropha H16.Appl Environ Microbiol. 2015 Mar;81(5):1847-58. doi: 10.1128/AEM.03791-14. Epub 2014 Dec 29. Appl Environ Microbiol. 2015. PMID: 25548058 Free PMC article.
-
Untargeted metabolomics analysis of Ralstonia eutropha during plant oil cultivations reveals the presence of a fucose salvage pathway.Sci Rep. 2021 Jul 12;11(1):14267. doi: 10.1038/s41598-021-93720-9. Sci Rep. 2021. PMID: 34253787 Free PMC article.
-
Versatile metabolic adaptations of Ralstonia eutropha H16 to a loss of PdhL, the E3 component of the pyruvate dehydrogenase complex.Appl Environ Microbiol. 2011 Apr;77(7):2254-63. doi: 10.1128/AEM.02360-10. Epub 2011 Feb 4. Appl Environ Microbiol. 2011. PMID: 21296938 Free PMC article.
-
PhaM is the physiological activator of poly(3-hydroxybutyrate) (PHB) synthase (PhaC1) in Ralstonia eutropha.Appl Environ Microbiol. 2014 Jan;80(2):555-63. doi: 10.1128/AEM.02935-13. Epub 2013 Nov 8. Appl Environ Microbiol. 2014. PMID: 24212577 Free PMC article.
-
Comparative Genomics of Marine Bacteria from a Historically Defined Plastic Biodegradation Consortium with the Capacity to Biodegrade Polyhydroxyalkanoates.Microorganisms. 2021 Jan 16;9(1):186. doi: 10.3390/microorganisms9010186. Microorganisms. 2021. PMID: 33467086 Free PMC article.
References
-
- Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. - PubMed
-
- Delafield, F. P., K. E. Cooksey, and M. Doudoroff. 1965. β-Hydroxybutyric dehydrogenase and dimer hydrolase of Pseudomonas lemoignei. J. Biol. Chem. 240:4023-4028. - PubMed
-
- Gao, D., A. Maehara, T. Yamane, and S. Ueda. 2001. Identification of the intracellular polyhydroxyalkanoate depolymerase gene of Paracoccus denitrificans and some properties of the gene product. FEMS Microbiol. Lett. 196:159-164. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

