Bacillus subtilis phosphorylated PhoP: direct activation of the E(sigma)A- and repression of the E(sigma)E-responsive phoB-PS+V promoters during pho response
- PMID: 16030210
- PMCID: PMC1196004
- DOI: 10.1128/JB.187.15.5166-5178.2005
Bacillus subtilis phosphorylated PhoP: direct activation of the E(sigma)A- and repression of the E(sigma)E-responsive phoB-PS+V promoters during pho response
Abstract
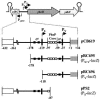
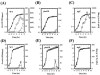
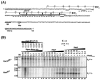
The phoB gene of Bacillus subtilis encodes an alkaline phosphatase (PhoB, formerly alkaline phosphatase III) that is expressed from separate promoters during phosphate deprivation in a PhoP-PhoR-dependent manner and at stage two of sporulation under phosphate-sufficient conditions independent of PhoP-PhoR. Isogenic strains containing either the complete phoB promoter or individual phoB promoter fusions were used to assess expression from each promoter under both induction conditions. The phoB promoter responsible for expression during sporulation, phoB-P(S), was expressed in a wild-type strain during phosphate deprivation, but induction occurred >3 h later than induction of Pho regulon genes and the levels were approximately 50-fold lower than that observed for the PhoPR-dependent promoter, phoB-P(V). E(sigma)E was necessary and sufficient for P(S) expression in vitro. P(S) expression in a phoPR mutant strain was delayed 2 to 3 h compared to the expression in a wild-type strain, suggesting that expression or activation of sigma(E) is delayed in a phoPR mutant under phosphate-deficient conditions, an observation consistent with a role for PhoPR in spore development under these conditions. Phosphorylated PhoP (PhoP approximately P) repressed P(S) in vitro via direct binding to the promoter, the first example of an E(sigma)E-responsive promoter that is repressed by PhoP approximately P. Whereas either PhoP or PhoP approximately P in the presence of E(sigma)A was sufficient to stimulate transcription from the phoB-P(V) promoter in vitro, roughly 10- and 17-fold-higher concentrations of PhoP than of PhoP approximately P were required for P(V) promoter activation and maximal promoter activity, respectively. The promoter for a second gene in the Pho regulon, ykoL, was also activated by elevated concentrations of unphosphorylated PhoP in vitro. However, because no Pho regulon gene expression was observed in vivo during P(i)-replete growth and PhoP concentrations increased only threefold in vivo during phoPR autoinduction, a role for unphosphorylated PhoP in Pho regulon activation in vivo is not likely.
Figures

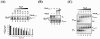


References
-
- Ames, B. N., and D. T. Dubin. 1960. The role of polyamines in the neutralization of bacteriophage deoxyribonucleic acid. J. Biol. Chem. 235:769-775. - PubMed
-
- Birkey, S. M., W. Liu, X. Zhang, M. F. Duggan, and F. M. Hulett. 1998. Pho signal transduction network reveals direct transcriptional regulation of one two-component system by another two-component regulator: Bacillus subtilis PhoP directly regulates production of ResD. Mol. Microbiol. 30:943-953. - PubMed
-
- Birkey, S. M., G. Sun, P. J. Piggot, and F. M. Hulett. 1994. A pho regulon promoter induced under sporulation conditions. Gene 147:95-100. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

