Mosaic nature of the wolbachia surface protein
- PMID: 16030235
- PMCID: PMC1196003
- DOI: 10.1128/JB.187.15.5406-5418.2005
Mosaic nature of the wolbachia surface protein
Abstract
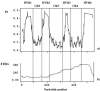
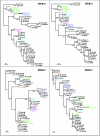
Lateral gene transfer and recombination play important roles in the evolution of many parasitic bacteria. Here we investigate intragenic recombination in Wolbachia bacteria, considered among the most abundant intracellular bacteria on earth. We conduct a detailed analysis of the patterns of variation and recombination within the Wolbachia surface protein, utilizing an extensive set of published and new sequences from five main supergroups of Wolbachia. Analysis of nucleotide and amino acid sequence variations confirms four hypervariable regions (HVRs), separated by regions under strong conservation. Comparison of shared polymorphisms reveals a complex mosaic structure of the gene, characterized by a clear intragenic recombining of segments among several distinct strains, whose major recombination effect is shuffling of a relatively conserved set of amino acid motifs within each of the four HVRs. Exchanges occurred both within and between the arthropod supergroups. Analyses based on phylogenetic methods and a specific recombination detection program (MAXCHI) significantly support this complex partitioning of the gene, indicating a chimeric origin of wsp. Although wsp has been widely used to define macro- and microtaxonomy among Wolbachia strains, these results clearly show that it is not suitable for this purpose. The role of wsp in bacterium-host interactions is currently unknown, but results presented here indicate that exchanges of HVR motifs are favored by natural selection. Identifying host proteins that interact with wsp variants should help reveal how these widespread bacterial parasites affect and evolve in response to the cellular environments of their invertebrate hosts.
Figures


References
-
- Baldo, L., J. D. Bartos, J. H. Werren, C. Bazzocchi, M. Casiraghi, and S. Panelli. 2002. Different rates of nucleotide substitutions in Wolbachia endosymbionts of arthropods and nematodes: arms race or host shifts? Parasitologia 44:179-187. - PubMed
-
- Bandi, C., T. J. C. Anderson, C. Genchi, and M. Blaxter. 2001. The Wolbachia endosymbionts of filarial nematodes, p. 25-43. In M. W. Kennedy and W. Harnett (ed.), Parasitic nematodes. CAB International, Wallingford, Oxon, United Kingdom.
-
- Bandi, C., A. J. Trees, and N. W. Brattig. 2001. Wolbachia in filarial nematodes: evolutionary aspects and implications for the pathogenesis and treatment of filarial diseases. Vet. Parasitol. 98:215-238. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

