Haem repression of the housekeeping 5-aminolaevulinic acid synthase gene in the hepatoma cell line LMH
- PMID: 16033334
- PMCID: PMC1317676
- DOI: 10.1042/BJ20050354
Haem repression of the housekeeping 5-aminolaevulinic acid synthase gene in the hepatoma cell line LMH
Abstract
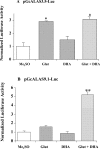
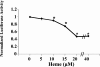
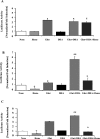
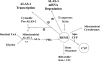
Haem is essential for the health and function of nearly all cells. 5-Aminolaevulinic acid synthase-1 (ALAS-1) catalyses the first and rate-controlling step of haem biosynthesis. ALAS-1 is repressed by haem and is induced strongly by lipophilic drugs that also induce CYP (cytochrome P450) proteins. We investigated the effects on the avian ALAS-1 gene promoter of a phenobarbital-like chemical, Glut (glutethimide), and a haem synthesis inhibitor, DHA (4,6-dioxoheptanoic acid), using a reporter gene assay in transiently transfected LMH (Leghorn male hepatoma) hepatoma cells. A 9.1 kb cALAS-1 (chicken ALAS-1) promoter-luciferase-reporter construct, was poorly induced by Glut and not by DHA alone, but was synergistically induced by the combination. In contrast, a 3.5 kb promoter ALAS-1 construct was induced by Glut alone, without any further effect of DHA. In addition, exogenous haem (20 microM) repressed the basal and Glut- and DHA-induced activity of luciferase reporter constructs containing 9.1 and 6.3 kb of ALAS-1 5'-flanking region but not the construct containing the first 3.5 kb of promoter sequence. This effect of haem was subsequently shown to be dependent on the -6.3 to -3.5 kb region of the 5'-flanking region of cALAS-1 and requires the native orientation of the region. Two deletion constructs of this approx. 2.8 kb haem-repressive region (1.7 and 1.1 kb constructs) retained haem-dependent repression of basal and drug inductions, suggesting that more than one cis-acting elements are responsible for this haem-dependent repression of ALAS-1. These results demonstrate that there are regulatory regions in the 5'-flanking region of the cALAS-1 gene that respond to haem and provide a basis for further investigations of the molecular mechanisms by which haem down-regulates expression of the ALAS-1 gene.
Figures



References
-
- Ades I. Z. Heme production in animal tissues: the regulation of biogenesis of delta-aminolevulinate synthase. Int. J. Biochem. 1990;22:565–578. - PubMed
-
- Bonkovsky H. L. Porphyrin and heme metabolism and the porphyrias. In: Zakim D., Boyer T., editors. Hepatology: A Textbook of Liver Disease. Philadelphia, PA: W.B. Saunders; 1990. pp. 378–424.
-
- May B. K., Dogra S. C., Sadlon T. J., Bhasker C. R., Cox T. C., Bottomley S. S. Molecular regulation of heme biosynthesis in higher vertebrates. Prog. Nucleic Acid Res. Mol. Biol. 1995;51:1–51. - PubMed
-
- Kappas A., Sassa S., Galbraith R. A., Nordmann Y. The porphyrias. In: Scriver C. R., Beaudet A. L., Sly W. S., Valle D., editors. The Metabolic Basis of Inherited Disease. New York: McGraw-Hill; 1995. pp. 2103–2160.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

