A computational approach for identifying pathogenicity islands in prokaryotic genomes
- PMID: 16033657
- PMCID: PMC1188055
- DOI: 10.1186/1471-2105-6-184
A computational approach for identifying pathogenicity islands in prokaryotic genomes
Abstract
Background: Pathogenicity islands (PAIs), distinct genomic segments of pathogens encoding virulence factors, represent a subgroup of genomic islands (GIs) that have been acquired by horizontal gene transfer event. Up to now, computational approaches for identifying PAIs have been focused on the detection of genomic regions which only differ from the rest of the genome in their base composition and codon usage. These approaches often lead to the identification of genomic islands, rather than PAIs.
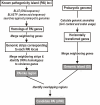
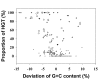
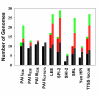
Results: We present a computational method for detecting potential PAIs in complete prokaryotic genomes by combining sequence similarities and abnormalities in genomic composition. We first collected 207 GenBank accessions containing either part or all of the reported PAI loci. In sequenced genomes, strips of PAI-homologs were defined based on the proximity of the homologs of genes in the same PAI accession. An algorithm reminiscent of sequence-assembly procedure was then devised to merge overlapping or adjacent genomic strips into a large genomic region. Among the defined genomic regions, PAI-like regions were identified by the presence of homolog(s) of virulence genes. Also, GIs were postulated by calculating G+C content anomalies and codon usage bias. Of 148 prokaryotic genomes examined, 23 pathogenic and 6 non-pathogenic bacteria contained 77 candidate PAIs that partly or entirely overlap GIs.
Conclusion: Supporting the validity of our method, included in the list of candidate PAIs were thirty four PAIs previously identified from genome sequencing papers. Furthermore, in some instances, our method was able to detect entire PAIs for those only partial sequences are available. Our method was proven to be an efficient method for demarcating the potential PAIs in our study. Also, the function(s) and origin(s) of a candidate PAI can be inferred by investigating the PAI queries comprising it. Identification and analysis of potential PAIs in prokaryotic genomes will broaden our knowledge on the structure and properties of PAIs and the evolution of bacterial pathogenesis.
Figures

Similar articles
-
PredictBias: a server for the identification of genomic and pathogenicity islands in prokaryotes.In Silico Biol. 2008;8(3-4):223-34. In Silico Biol. 2008. PMID: 19032158
-
Towards pathogenomics: a web-based resource for pathogenicity islands.Nucleic Acids Res. 2007 Jan;35(Database issue):D395-400. doi: 10.1093/nar/gkl790. Epub 2006 Nov 7. Nucleic Acids Res. 2007. PMID: 17090594 Free PMC article.
-
Pathogenicity islands: a molecular toolbox for bacterial virulence.Cell Microbiol. 2006 Nov;8(11):1707-19. doi: 10.1111/j.1462-5822.2006.00794.x. Epub 2006 Aug 24. Cell Microbiol. 2006. PMID: 16939533 Review.
-
[Plasticity of bacterial genomes: pathogenicity islands and the locus of enterocyte effacement (LEE)].Berl Munch Tierarztl Wochenschr. 2004 Mar-Apr;117(3-4):116-29. Berl Munch Tierarztl Wochenschr. 2004. PMID: 15046458 Review. German.
-
Impact of pathogenicity islands in bacterial diagnostics.APMIS. 2004 Nov-Dec;112(11-12):930-6. doi: 10.1111/j.1600-0463.2004.apm11211-1214.x. APMIS. 2004. PMID: 15638844 Review.
Cited by
-
Identification of prophages in bacterial genomes by dinucleotide relative abundance difference.PLoS One. 2007 Nov 21;2(11):e1193. doi: 10.1371/journal.pone.0001193. PLoS One. 2007. PMID: 18030328 Free PMC article.
-
High-density transcriptional initiation signals underline genomic islands in bacteria.PLoS One. 2012;7(3):e33759. doi: 10.1371/journal.pone.0033759. Epub 2012 Mar 20. PLoS One. 2012. PMID: 22448273 Free PMC article.
-
On detection and assessment of statistical significance of Genomic Islands.BMC Genomics. 2008 Apr 1;9:150. doi: 10.1186/1471-2164-9-150. BMC Genomics. 2008. PMID: 18380895 Free PMC article.
-
Distribution of pathogenicity island (PAI) markers and phylogenetic groups in diarrheagenic and commensal Escherichia coli from young children.Gastroenterol Hepatol Bed Bench. 2016 Fall;9(4):316-324. Gastroenterol Hepatol Bed Bench. 2016. PMID: 27895858 Free PMC article.
-
IGIPT - Integrated genomic island prediction tool.Bioinformation. 2011;7(6):307-10. doi: 10.6026/007/97320630007307. Epub 2011 Nov 20. Bioinformation. 2011. PMID: 22355227 Free PMC article.
References
-
- Hacker J, Kaper JB. Pathogenicity islands and the evolution of pathogenic microbes. Berlin , Springer-Verlag; 2002.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

