Alterations in the alpha2 isoform of Na,K-ATPase associated with familial hemiplegic migraine type 2
- PMID: 16037212
- PMCID: PMC1178013
- DOI: 10.1073/pnas.0504323102
Alterations in the alpha2 isoform of Na,K-ATPase associated with familial hemiplegic migraine type 2
Abstract
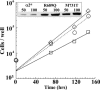
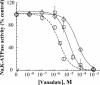
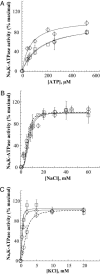
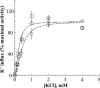
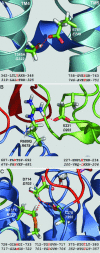
A number of missense mutations in the Na,K-ATPase alpha2 catalytic subunit have been identified in familial hemiplegic migraine with aura. Two alleles (L764P and W887R) showed loss-of-function, whereas a third (T345A) is fully functional but with altered Na,K-ATPase kinetics. This study describes two additional mutants, R689Q and M731T, originally identified by Vanmolkot et al. [Vanmolkot, K. R., et al. (2003) Ann. Neurol. 54, 360-366], which we show here to also be functional and kinetically altered. Both mutants have reduced catalytic turnover and increased apparent affinity for extracellular K(+). For both R689Q and M731T, sensitivity to vanadate inhibition is decreased, suggesting that the steady-state E(1) <==> E(2) poise of the enzyme is shifted toward E(1). Whereas the K'(ATP) is not affected by the R689Q replacement, the M731T mutant has an increase in apparent affinity for ATP. Analysis of the structural changes effected by T345A, R689Q, and M731T mutations, based on homologous replacements in the known crystal structure of the sarcoplasmic reticulum Ca-ATPase, provides insights into the molecular bases for the kinetic alterations. It is suggested that the disease phenotype is the consequence of lowered molecular activity of the alpha2 pump isoform due to either decreased K(+) affinity (T345A) or catalytic turnover (R689Q and M731T), thus causing a delay in extracellular K(+) clearance and/or altered localized Ca(2+) handling/signaling secondary to reduced activity in colocalized Na(+)/Ca(2+) exchange.
Figures
References
-
- Kaplan, J. H. (2002) Annu. Rev. Biochem. 71, 511-535. - PubMed
-
- Jorgensen, P. L., Hakansson, K. O. H. & Karlish, S. J. D. (2003) Ann. Rev. Physiol. 65, 817-849. - PubMed
-
- Sweadner, K. J. (1989) Biochim. Biophys. Acta 988, 185-220. - PubMed
-
- Moseley, A. E., Dean, W. L. & Delamere, N. A. (1996) Invest. Ophthalmol. Visual Sci. 37, 1502-1508. - PubMed
-
- Orlowski, J. & Lingrel, J. B. (1988) J. Biol. Chem. 263, 10436-10442. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

