Nuclear export and cytoplasmic processing of precursors to the 40S ribosomal subunits in mammalian cells
- PMID: 16037817
- PMCID: PMC1187937
- DOI: 10.1038/sj.emboj.7600752
Nuclear export and cytoplasmic processing of precursors to the 40S ribosomal subunits in mammalian cells
Abstract
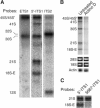
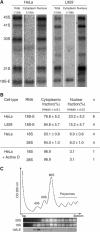
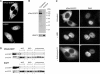
It is generally assumed that, in mammalian cells, preribosomal RNAs are entirely processed before nuclear exit. Here, we show that pre-40S particles exported to the cytoplasm in HeLa cells contain 18S rRNA extended at the 3' end with 20-30 nucleotides of the internal transcribed spacer 1. Maturation of this pre-18S rRNA (which we named 18S-E) involves a cytoplasmic protein, the human homolog of the yeast kinase Rio2p, and appears to be required for the translation competence of the 40S subunit. By tracking the nuclear exit of this precursor, we have identified the ribosomal protein Rps15 as a determinant of preribosomal nuclear export in human cells. Interestingly, inhibition of exportin Crm1/Xpo1 with leptomycin B strongly alters processing of the 5'-external transcribed spacer, upstream of nuclear export, and reveals a new cleavage site in this transcribed spacer. Completion of the maturation of the 18S rRNA in the cytoplasm, a feature thought to be unique to yeast, may prevent pre-40S particles from initiating translation with pre-mRNAs in eukaryotic cells. It also allows new strategies for the study of preribosomal transport in mammalian cells.
Figures


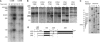
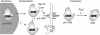
References
-
- Boulon S, Verheggen C, Jady BE, Girard C, Pescia C, Paul C, Ospina JK, Kiss T, Matera AG, Bordonne R, Bertrand E (2004) PHAX and CRM1 are required sequentially to transport U3 snoRNA to nucleoli. Mol Cell 16: 777–787 - PubMed
-
- Eichler DC, Craig N (1994) Processing of eukaryotic ribosomal RNA. Prog Nucleic Acid Res Mol Biol 49: 197–239 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

