Four new type I restriction enzymes identified in Escherichia coli clinical isolates
- PMID: 16040596
- PMCID: PMC1178010
- DOI: 10.1093/nar/gni114
Four new type I restriction enzymes identified in Escherichia coli clinical isolates
Abstract
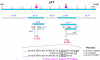
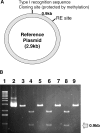
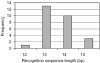
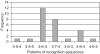
Using a plasmid transformation method and the RM search computer program, four type I restriction enzymes with new recognition sites and two isoschizomers (EcoBI and Eco377I) were identified in a collection of clinical Escherichia coli isolates. These new enzymes were designated Eco394I, Eco826I, Eco851I and Eco912I. Their recognition sequences were determined to be GAC(5N)RTAAY, GCA(6N)CTGA, GTCA(6N)TGAY and CAC(5N)TGGC, respectively. A methylation sensitivity assay, using various synthetic oligonucleotides, was used to identify the adenines that prevent cleavage when methylated (underlined). These results suggest that type I enzymes are abundant in E.coli and many other bacteria, as has been inferred from bacterial genome sequencing projects.
Figures
References
-
- Bullas L.R., Colson C., Van Pel A. DNA restriction and modification systems in Salmonella. SQ, a new system derived by recombination between the SB system of Salmonella typhimurium and the SP system of Salmonella potsdam. J. Gen. Microbiol. 1976;95:166–172. - PubMed
-
- Murray N.E. 2001 Fred Griffith review lecture. Immigration control of DNA in bacteria: self versus non-self. Microbiology. 2002;148:3–20. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

