Arabidopsis peroxin 16 coexists at steady state in peroxisomes and endoplasmic reticulum
- PMID: 16040658
- PMCID: PMC1183388
- DOI: 10.1104/pp.105.061291
Arabidopsis peroxin 16 coexists at steady state in peroxisomes and endoplasmic reticulum
Abstract
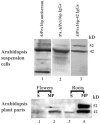
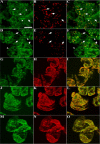
Homologs of peroxin 16 genes (PEX16) have been identified only in Yarrowia lipolytica, humans (Homo sapiens), and Arabidopsis (Arabidopsis thaliana). The Arabidopsis gene (AtPEX16), previously reported as the SSE1 gene, codes for a predicted 42-kD membrane peroxin protein (AtPex16p). Lin et al. (Y. Lin, J.E. Cluette-Brown, H.M. Goodman [2004] Plant Physiol 135: 814-827) reported that SSE1/AtPEX16 was essential for endoplasmic reticulum (ER)-dependent oil and protein body biogenesis in peroxisome-deficient maturing seeds and likely also was involved in peroxisomal biogenesis based on localization of stably expressed green fluorescent protein::AtPex16p in peroxisomes of Arabidopsis plants. In this study with Arabidopsis suspension-cultured cells, combined in vivo and in vitro experiments revealed a novel dual organelle localization and corresponding membrane association/topology of endogenous AtPex16p. Immunofluorescence microscopy with antigen affinity-purified IgGs showed an unambiguous, steady-state coexistence of AtPex16p in suspension cell peroxisomes and ER. AtPex16p also was observed in peroxisomes and ER of root and leaf cells. Cell fractionation experiments surprisingly revealed two immunorelated polypeptides, 42 kD (expected) and 52 kD (unexpected), in homogenates and microsome membrane pellets derived from roots, inflorescence, and suspension cells. Suc-gradient purifications confirmed the presence of both 42-kD and 52-kD polypeptides in isolated peroxisomes (isopycnic separation) and in rough ER vesicles (Mg2+ shifted). They were found peripherally associated with peroxisome and ER membranes but not as covalently bound subunits of AtPex16p. Both were mostly on the matrix side of peroxisomal membranes and unexpectedly mostly on the cytosolic side of ER membranes. In summary, AtPex16p is the only authentic plant peroxin homolog known to coexist at steady state within peroxisomes and ER; these data provide new insights in support of its ER-related, multifunctional roles in organelle biogenesis.
Figures









Similar articles
-
Arabidopsis peroxin 16 trafficks through the ER and an intermediate compartment to pre-existing peroxisomes via overlapping molecular targeting signals.J Exp Bot. 2007;58(7):1677-93. doi: 10.1093/jxb/erm018. Epub 2007 Apr 12. J Exp Bot. 2007. PMID: 17431024
-
Five Arabidopsis peroxin 11 homologs individually promote peroxisome elongation, duplication or aggregation.J Cell Sci. 2006 May 1;119(Pt 9):1961-72. doi: 10.1242/jcs.02904. J Cell Sci. 2006. PMID: 16636080
-
The Arabidopsis PEX12 gene is required for peroxisome biogenesis and is essential for development.Plant Physiol. 2005 Sep;139(1):231-9. doi: 10.1104/pp.105.066811. Epub 2005 Aug 19. Plant Physiol. 2005. PMID: 16113209 Free PMC article.
-
Peroxisome biogenesis, protein targeting mechanisms and PEX gene functions in plants.Biochim Biophys Acta. 2016 May;1863(5):850-62. doi: 10.1016/j.bbamcr.2015.09.027. Epub 2015 Sep 25. Biochim Biophys Acta. 2016. PMID: 26408938 Review.
-
PEX16: a multifaceted regulator of peroxisome biogenesis.Front Physiol. 2013 Sep 3;4:241. doi: 10.3389/fphys.2013.00241. Front Physiol. 2013. PMID: 24027535 Free PMC article. Review.
Cited by
-
Peroxisome biogenesis, membrane contact sites, and quality control.EMBO Rep. 2019 Jan;20(1):e46864. doi: 10.15252/embr.201846864. Epub 2018 Dec 10. EMBO Rep. 2019. PMID: 30530632 Free PMC article. Review.
-
Arabidopsis DAYU/ABERRANT PEROXISOME MORPHOLOGY9 is a key regulator of peroxisome biogenesis and plays critical roles during pollen maturation and germination in planta.Plant Cell. 2014 Feb;26(2):619-35. doi: 10.1105/tpc.113.121087. Epub 2014 Feb 7. Plant Cell. 2014. PMID: 24510720 Free PMC article.
-
De novo peroxisome biogenesis: Evolving concepts and conundrums.Biochim Biophys Acta. 2016 May;1863(5):892-901. doi: 10.1016/j.bbamcr.2015.09.014. Epub 2015 Sep 14. Biochim Biophys Acta. 2016. PMID: 26381541 Free PMC article. Review.
-
De novo peroxisome biogenesis in Penicillium chrysogenum is not dependent on the Pex11 family members or Pex16.PLoS One. 2012;7(4):e35490. doi: 10.1371/journal.pone.0035490. Epub 2012 Apr 19. PLoS One. 2012. PMID: 22536392 Free PMC article.
-
Localization of the tomato bushy stunt virus replication protein p33 reveals a peroxisome-to-endoplasmic reticulum sorting pathway.Plant Cell. 2005 Dec;17(12):3513-31. doi: 10.1105/tpc.105.036350. Epub 2005 Nov 11. Plant Cell. 2005. PMID: 16284309 Free PMC article.
References
-
- Baerends RJS, Faber KN, Kiel JAKW, Van der Klei IJ, Harder W, Veenhuis M (2000) Sorting and function of peroxisomal membrane proteins. FEMS Microbiol Rev 24: 291–301 - PubMed
-
- Baker A, Graham IA (2002) Plant Peroxisomes: Biochemistry, Cell Biology and Biotechnological Applications. Kluwer Academic Publishers, Dordrecht, The Netherlands
-
- Charlton W, Lopez-Huertas E (2002) PEX genes in plants and other organisms. In A Baker, I Graham, eds, Plant Peroxisomes. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 385–226
-
- Corpas FJ, Bunkelmann J, Trelease RN (1994) Identification and immunochemical characterization of a family of peroxisome membrane-proteins (Pmps) in oilseed glyoxysomes. Eur J Cell Biol 65: 280–290 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

