Exposure to rumen protozoa leads to enhancement of pathogenicity of and invasion by multiple-antibiotic-resistant Salmonella enterica bearing SGI1
- PMID: 16040979
- PMCID: PMC1201270
- DOI: 10.1128/IAI.73.8.4668-4675.2005
Exposure to rumen protozoa leads to enhancement of pathogenicity of and invasion by multiple-antibiotic-resistant Salmonella enterica bearing SGI1
Abstract
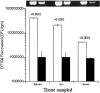
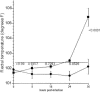
Multiple-antibiotic-resistant Salmonella enterica serotype Typhimurium is a food-borne pathogen that has been purported to be more virulent than antibiotic-sensitive counterparts. The paradigm for this multiresistant/hyperpathogenic phenotype is Salmonella enterica serotype Typhimurium phage type DT104 (DT104). The basis for the multiresistance in DT104 is related to an integron structure designated SGI1, but factors underlying hyperpathogenicity have not been completely identified. Since protozoa have been implicated in the alteration of virulence in Legionella and Mycobacterium spp., we attempted to assess the possibility that protozoa may contribute to the putative hypervirulence of DT104. Our study reveals that DT104 can be more invasive, as determined by a tissue culture invasion assay, after surviving within protozoa originating from the bovine rumen. The enhancement of invasion was correlated with hypervirulence in a bovine infection model in which we observed a more rapid progression of disease and a greater recovery rate for the pathogen. Fewer DT104 cells were recovered from tissues of infected animals when protozoa were lysed by preinfection chemical defaunation of the bovine or ovine rumen. The protozoan-mediated hypervirulence phenotype was observed only in DT104 and other Salmonella strains, including serovars Agona and Infantis, possessing SGI1.
Figures




References
-
- Boyd, D., G. Peters, A. Cloeckaert, K. Boumedine, E. Chaslus-Dancla, H. Imberechts, and M. Mulvey. 2001. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 183:5725-5732. - PMC - PubMed
-
- Carlson, S. A., L. F. Bolton, C. E. Briggs, H. S. Hurd, V. K. Sharma, P. Fedorka-Cray, and B. D. Jones. 1999. Detection of Salmonella typhimurium DT104 using multiplex and fluorogenic PCR. Mol. Cell. Probes 13:213-222. - PubMed
-
- Carlson, S. A., M. Browning, K. E. Ferris, and B. D. Jones. 2000. Identification of diminished tissue culture invasiveness among multiple antibiotic resistant Salmonella typhimurium DT104. Microb. Pathog. 28:37-44. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

