Comparison of Helicobacter pylori virulence gene expression in vitro and in the Rhesus macaque
- PMID: 16041003
- PMCID: PMC1201232
- DOI: 10.1128/IAI.73.8.4895-4904.2005
Comparison of Helicobacter pylori virulence gene expression in vitro and in the Rhesus macaque
Abstract
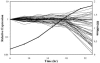
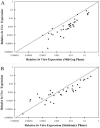
We used a quantitative real-time reverse transcriptase PCR assay to measure the transcript abundance of 46 known and putative Helicobacter pylori virulence genes, including 24 genes on the Cag pathogenicity island. The expression profile of H. pylori cells grown in vitro was also compared to expression in vivo after experimental infection of rhesus macaques. Transcript abundance in vitro (mid-log phase) ranged from about 0.004 (feoB and hpaA) to 20 (ureAB, napA, and cag25) copies/cell. Expression of most genes was repressed during the transition from logarithmic- to stationary-phase growth, but several well-characterized H. pylori virulence genes (katA, napA, vacA, and cagA) were induced. Comparison of results in the rhesus macaque with similar data from humans showed a strong correlation (r = 0.89). The relative in vivo expression in the rhesus monkey was highly correlated with in vitro expression during mid-log (r = 0.89)- and stationary (r = 0.88)-phase growth. Transcript abundance was on average three- to fourfold reduced in vivo compared to in vitro during mid-log phase. However, when compared to stationary phase, increased expression in vivo was observed for 6 of 7 genes on a contiguous portion of the pathogenicity island, several of which are thought to encode the H. pylori type IV structural pilus and its accessory proteins. These results suggest the possibility that some genes encoding the H. pylori type IV structural pilus and accessory proteins may form an operon that is induced during growth in vivo.
Figures




References
-
- Akada, J. K., M. Shirai, H. Takeuchi, M. Tsuda, and T. Nakazawa. 2000. Identification of the urease operon in Helicobacter pylori and its control by mRNA decay in response to pH. Mol. Microbiol. 36:1071-1084. - PubMed
-
- Akopyants, N. S., S. W. Clifton, D. Kersulyte, J. E. Crabtree, B. E. Youree, C. A. Reece, N. O. Bukanov, E. S. Drazek, B. A. Roe, and D. E. Berg. 1998. Analyses of the cag pathogenicity island of Helicobacter pylori. Mol. Microbiol. 28:37-53. - PubMed
-
- Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. - PubMed
-
- Aspholm-Hurtig, M., G. Dailide, M. Lahmann, A. Kalia, D. Ilver, N. Roche, S. Vikstrom, R. Sjostrom, S. Linden, A. Backstrom, C. Lundberg, A. Arnqvist, J. Mahdavi, U. J. Nilsson, B. Velapatino, R. H. Gilman, M. Gerhard, T. Alarcon, M. Lopez-Brea, T. Nakazawa, J. G. Fox, P. Correa, M. G. Dominguez-Bello, G. I. Perez-Perez, M. J. Blaser, S. Normark, I. Carlstedt, S. Oscarson, S. Teneberg, D. E. Berg, and T. Boren. 2004. Functional adaptation of BabA, the H. pylori ABO blood group antigen binding adhesin. Science 305:519-522. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

