The role of ribosome recycling factor in dissociation of 70S ribosomes into subunits
- PMID: 16043510
- PMCID: PMC1370814
- DOI: 10.1261/rna.2520405
The role of ribosome recycling factor in dissociation of 70S ribosomes into subunits
Abstract
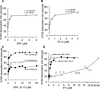
Protein synthesis is initiated on ribosomal subunits. However, it is not known how 70S ribosomes are dissociated into small and large subunits. Here we show that 70S ribosomes, as well as the model post-termination complexes, are dissociated into stable subunits by cooperative action of three translation factors: ribosome recycling factor (RRF), elongation factor G (EF-G), and initiation factor 3 (IF3). The subunit dissociation is stable enough to be detected by conventional sucrose density gradient centrifugation (SDGC). GTP, but not nonhydrolyzable GTP analog, is essential in this process. We found that RRF and EF-G alone transiently dissociate 70S ribosomes. However, the transient dissociation cannot be detected by SDGC. IF3 stabilizes the dissociation by binding to the transiently formed 30S subunits, preventing re-association back to 70S ribosomes. The three-factor-dependent stable dissociation of ribosomes into subunits completes the ribosome cycle and the resulting subunits are ready for the next round of translation.
Figures




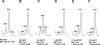

References
-
- Agrawal, R.K. and Burma, D.P. 1996. Sites of ribosomal RNAs involved in the subunit association of tight and loose couple ribosomes. J. Biol. Chem. 271: 21285–21291. - PubMed
-
- Andersen, L.D., Moreno, J.M.D., Clark, B.F.C., Mortensen, K.K., and Sperling-Petersen, H.U. 1999. Immunochemical determination of cellular content of translation release factor RF4 in Escherichia coli. IUBMB Life 48: 283–286. - PubMed
-
- Bodley, J.W., Zieve, F.J., Lin, L., and Zieve, S.T. 1970. Studies on translocation III. Conditions necessary for the formation and detection of a stable ribosome-G factor-guanosine diphosphate complex in the presence of fusidic acid. J. Biol. Chem. 245: 5656–5661. - PubMed
-
- Caldas, T., Binet, E., Bouloc, P., and Richarme, G. 2000. Translational defects of Escherichia coli mutants deficient in the Um(2552) 23S ribosomal RNA methyltransferase RrmJ/FTSJ. Biochem. Biophys. Res. Commun. 271: 714–718. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
