The structure of phospholamban pentamer reveals a channel-like architecture in membranes
- PMID: 16043693
- PMCID: PMC1182456
- DOI: 10.1073/pnas.0504920102
The structure of phospholamban pentamer reveals a channel-like architecture in membranes
Abstract
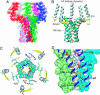
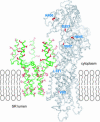
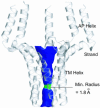
Contraction and relaxation of heart muscle cells is regulated by cycling of calcium between cytoplasm and sarcoplasmic reticulum. Human phospholamban (PLN), expressed in the sarcoplasmic reticulum membrane as a 30-kDa homopentamer, controls cellular calcium levels by a mechanism that depends on its phosphorylation. Since PLN was discovered approximately 30 years ago, extensive studies have aimed to explain how it influences calcium pumps and to determine whether it acts as an ion channel. We have determined by solution NMR methods the atomic resolution structure of an unphosphorylated PLN pentamer in dodecylphosphocholine micelles. The unusual bellflower-like assembly is held together by leucine/isoleucine zipper motifs along the membrane-spanning helices. The structure reveals a channel-forming architecture that could allow passage of small ions. The central pore gradually widens toward the cytoplasmic end as the transmembrane helices twist around each other and bend outward. The dynamic N-terminal amphipathic helices point away from the membrane, perhaps facilitating recognition and inhibition of the calcium pump.
Figures
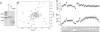

References
-
- MacLennan, D. H. & Kranias, E. G. (2003) Nat. Rev. Mol. Cell. Biol. 4, 566-577. - PubMed
-
- Reddy, L. G., Jones, L. R. & Thomas, D. D. (1999) Biochemistry 38, 3954-3962. - PubMed
-
- Kimura, Y., Kurzydlowski, K., Tada, M. & MacLennan, D. H. (1997) J. Biol. Chem. 272, 15061-15064. - PubMed
-
- Kovacs, R. J., Nelson, M. T., Simmerman, H. K. & Jones, L. R. (1988) J. Biol. Chem. 263, 18364-18368. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

