Diffuse large B-cell lymphoma subgroups have distinct genetic profiles that influence tumor biology and improve gene-expression-based survival prediction
- PMID: 16046532
- PMCID: PMC1895326
- DOI: 10.1182/blood-2005-04-1399
Diffuse large B-cell lymphoma subgroups have distinct genetic profiles that influence tumor biology and improve gene-expression-based survival prediction
Abstract
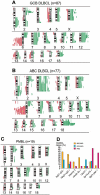
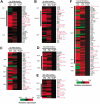
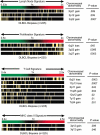
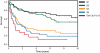
Gene-expression profiling has identified 3 major subgroups of diffuse large B-cell lymphoma (DLBCL): germinal center B-cell-like (GCB), activated B-cell-like (ABC), and primary mediastinal DLBCL (PMBCL). Using comparative genomic hybridization (CGH), we investigated the genetic alterations of 224 cases of untreated DLBCL (87 GCB-DLBCL, 77 ABC-DLBCL, 19 PMBCL, and 41 unclassified DLBCL) previously characterized by gene-expression profiling. The DLBCL subgroups differed significantly in the frequency of particular chromosomal aberrations. ABC-DLBCL had frequent trisomy 3, gains of 3q and 18q21-q22, and losses of 6q21-q22, whereas GCB-DLBCL had frequent gains of 12q12, and PMBCL had gains of 9p21-pter and 2p14-p16. Parallel analysis of CGH alterations, locus-specific gene-expression profiles, and global gene-expression signatures revealed that DNA amplifications and gains had a substantial impact on the expression of genes in the involved chromosomal regions, and some genes were overexpressed in a DLBCL subgroup-specific fashion. Unexpectedly, specific chromosomal alterations were associated with significant changes in gene-expression signatures that reflect various aspects of lymphoma cell biology as well as the host response to the lymphoma. In addition, gains involving the chromosomal region 3p11-p12 provided prognostic information that was statistically independent of the previously defined gene-expression-based survival model, thereby improving its predictive power.
Figures
References
-
- The Non-Hodgkin's Lymphoma Classification Project. A clinical evaluation of the International Lymphoma Study Group classification of non-Hodgkin's lymphoma. Blood. 1997;89: 3909-3918. - PubMed
-
- Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403: 503-511. - PubMed
-
- Rosenwald A, Wright G, Chan WC, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346: 1937-1947. - PubMed
-
- Savage KJ, Monti S, Kutok JL, et al. The molecular signature of mediastinal large B-cell lymphoma differs from that of other diffuse large B-cell lymphomas and shares features with classical Hodgkin lymphoma. Blood. 2003;102: 3871-3879. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

