Remodeling protein complexes: insights from the AAA+ unfoldase ClpX and Mu transposase
- PMID: 16046622
- PMCID: PMC2279306
- DOI: 10.1110/ps.051417505
Remodeling protein complexes: insights from the AAA+ unfoldase ClpX and Mu transposase
Abstract
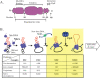
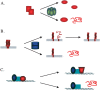
Multiprotein complexes in the cell are dynamic entities that are constantly undergoing changes in subunit composition and conformation to carry out their functions. The protein-DNA complex that promotes recombination of the bacteriophage Mu is a prime example of a complex that must undergo specific changes to carry out its function. The Clp/Hsp100 family of AAA+ ATPases plays a critical role in mediating such changes. The Clp/Hsp100 unfolding enzymes have been extensively studied for the roles they play in protein degradation. However, degradation is not the only fate for proteins that come in contact with the ATP-dependent unfolding enzymes. The Clp/Hsp100 enzymes induce structural changes in their substrates. These structural changes, which we refer to as "remodeling", ultimately change the biological activity of the substrate. These biological changes include activation, inactivation (not associated with degradation), and relocation within the cell. Analysis of the interaction between Escherichia coli ClpX unfoldase and the Mu recombination complex, has provided molecular insight into the mechanisms of protein remodeling. We discuss the key mechanistic features of the remodeling reactions promoted by ClpX and possible implications of these findings for other biological reactions.
Figures



References
-
- Aldaz, H., Schuster, E., and Baker, T.A. 1996. The interwoven architecture of the Mu transposase couples DNA synapsis to catalysis. Cell 85 257–269. - PubMed
-
- Baker, T.A. and Mizuuchi, K. 1992. DNA-promoted assembly of the active tetramer of the Mu transposase. Genes & Dev. 6 2221–2232. - PubMed
-
- Bochtler, M., Hartmann, C., Song, H.K., Bourenkov, G.P., Bartunik, H.D., and Huber, R. 2000. The structures of HsIU and the ATP-dependent protease HsIU-HsIV. Nature 403 800–805. - PubMed
-
- Burton, B.M. and Baker, T.A. 2003. Mu transpososome architecture ensures that unfolding by ClpX or proteolysis by ClpXP remodels but does not destroy the complex. Chem. Biol. 10 463–72. - PubMed
-
- Burton, B.M., Williams, T.L., and Baker, T.A. 2001. ClpX-mediated remodeling of mu transpososomes: Selective unfolding of subunits destabilizes the entire complex. Mol. Cell 8 449–454. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

