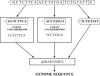
Genome-wide selection of unique and valid oligonucleotides
- PMID: 16049019
- PMCID: PMC1180749
- DOI: 10.1093/nar/gni110
Genome-wide selection of unique and valid oligonucleotides
Abstract
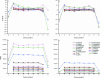
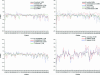
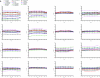
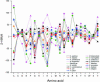
Functional genomics methods are used to investigate the huge amount of information contained in genomes. Numerous experimental methods rely on the use of oligo- or polynucleotides. Nucleotide strand hybridization forms the underlying principle for these methods. For all these techniques, the probes should be unique for analyzed genes. In addition to being unique for the studied genes, the probes should fulfill a large number of criteria to be usable and valid. The criteria include for example, avoidance of self-annealing, suitable melting temperature and nucleotide composition. We developed a method for searching unique and valid oligonucleotides or probes for genes so that there is not even a similar (approximate) occurrence in any other location of the whole genome. By using probe size 25, we analyzed 17 complete genomes representing a wide range of both prokaryotic and eukaryotic organisms. More than 92% of all the genes in the investigated genomes contained valid oligonucleotides. Extensive statistical tests were performed to characterize the properties of unique and valid oligonucleotides. Unique and valid oligonucleotides were relatively evenly distributed in genes except for the beginning and end, which were somewhat overrepresented. The flanking regions in eukaryotes were clearly underrepresented among suitable oligonucleotides. In addition to distributions within genes, the effects on codon and amino acid usage were also studied.
Figures




Similar articles
-
The computational detection of functional nucleotide sequence motifs in the coding regions of organisms.Exp Biol Med (Maywood). 2008 Jun;233(6):665-73. doi: 10.3181/0704-MR-97. Epub 2008 Apr 11. Exp Biol Med (Maywood). 2008. PMID: 18408149 Review.
-
ISSCOR: Intragenic, Stochastic Synonymous Codon Occurrence Replacement--a new method for an alignment-free genome sequence analysis.C R Biol. 2009 Apr;332(4):336-50. doi: 10.1016/j.crvi.2008.11.008. Epub 2009 Feb 3. C R Biol. 2009. PMID: 19304264
-
Where are we in genomics?J Physiol Pharmacol. 2005 Jun;56 Suppl 3:37-70. J Physiol Pharmacol. 2005. PMID: 16077195 Review.
-
Genetic robustness and selection at the protein level for synonymous codons.J Evol Biol. 2006 Mar;19(2):353-65. doi: 10.1111/j.1420-9101.2005.01029.x. J Evol Biol. 2006. PMID: 16599911
-
Unique genes in plants: specificities and conserved features throughout evolution.BMC Evol Biol. 2008 Oct 10;8:280. doi: 10.1186/1471-2148-8-280. BMC Evol Biol. 2008. PMID: 18847470 Free PMC article.
Cited by
-
Genome-wide identification of specific oligonucleotides using artificial neural network and computational genomic analysis.BMC Bioinformatics. 2007 May 22;8:164. doi: 10.1186/1471-2105-8-164. BMC Bioinformatics. 2007. PMID: 17518996 Free PMC article.
-
PrimerStation: a highly specific multiplex genomic PCR primer design server for the human genome.Nucleic Acids Res. 2006 Jul 1;34(Web Server issue):W665-9. doi: 10.1093/nar/gkl297. Nucleic Acids Res. 2006. PMID: 16845094 Free PMC article.
-
A Java-based tool for the design of classification microarrays.BMC Bioinformatics. 2008 Aug 4;9:328. doi: 10.1186/1471-2105-9-328. BMC Bioinformatics. 2008. PMID: 18680597 Free PMC article.
References
-
- Hillier L., Green P. OSP: a computer program for choosing PCR and DNA sequencing primers. PCR Methods Appl. 1991;1:124–128. - PubMed
-
- Cutichia A., Arnold J., Timberlake W.E. PCAP: probe choice and analysis package—a set of programs to aid in choosing synthetic oligomers for contig mapping. Comput. Appl. Biosci. 1993;9:201–203. - PubMed
-
- Li P., Kupfer K.C., Davies C.J., Burbee D., Evans G.A., Garner H.R. PRIMO: a primer design program that applies base quality statistics for automated large-scale DNA sequencing. Genomics. 1997;40:476–485. - PubMed
-
- Mecklenburg M. Design of high-annealing-temperature primers for PCR and development of a versatile low-copy-number amplification protocol. Adv. Mol. Cell Biol. 1997;15B:473–490.

