Induction of transcription within chromosomal DNA loops flanked by MAR elements causes an association of loop DNA with the nuclear matrix
- PMID: 16049024
- PMCID: PMC1180747
- DOI: 10.1093/nar/gki733
Induction of transcription within chromosomal DNA loops flanked by MAR elements causes an association of loop DNA with the nuclear matrix
Abstract
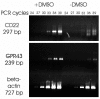
The spatial organization of an approximately 170 kb region of human chromosome 19, including CD22 and GPR40-GPR43 genes, was studied using in situ hybridization of a set of cosmid and PAC probes with nuclear halos prepared from proliferating and differentiated HL60 cells. The whole region under study was found to be looped out into the nuclear halo in proliferating cells. It is likely that the loop observed was attached to the nuclear matrix via MAR elements present at the flanks of the area under study. Upon dimethyl sulfoxide-induced differentiation of the cells the looped fragment became associated with the nuclear matrix. This change in the spatial organization correlated with the activation of transcription of at least two (CD22 and GPR43) genes present within the loop. The data obtained are discussed in the framework of the hypothesis postulating that the spatial organization of chromosomal DNA is maintained via constitutive (basic) and facultative (transcription-related) interactions of the latter with the nuclear matrix.
Figures



References
-
- Paulson J.R., Laemmli U.K. The structure of histone-depleted metaphase chromosomes. Cell. 1977;12:817–828. - PubMed
-
- Hancock R., Hughes M.E. Organization of DNA in the eukaryotic nucleus. Biol. Cell. 1982;44:201–212.
-
- Adolph K.W., Chang S.M., Laemmli U.K. Role of nonhistone proteins in metaphase chromosomes structure. Cell. 1977;12:805–816. - PubMed
-
- Buongiorno-Nardelli M., Gioacchino M., Carri M.T., Marilley M. A relationship between replicon size and supercoiled loop domains in the eukaryotic genome. Nature. 1982;298:100–102. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

