Genes that mediate breast cancer metastasis to lung
- PMID: 16049480
- PMCID: PMC1283098
- DOI: 10.1038/nature03799
Genes that mediate breast cancer metastasis to lung
Abstract
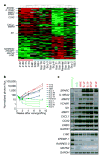
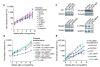
By means of in vivo selection, transcriptomic analysis, functional verification and clinical validation, here we identify a set of genes that marks and mediates breast cancer metastasis to the lungs. Some of these genes serve dual functions, providing growth advantages both in the primary tumour and in the lung microenvironment. Others contribute to aggressive growth selectively in the lung. Many encode extracellular proteins and are of previously unknown relevance to cancer metastasis.
Figures



References
-
- Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nature Rev Cancer. 2002;2:563–572. - PubMed
-
- Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nature Rev Cancer. 2003;3:453–458. - PubMed
-
- Yokota J. Tumor progression and metastasis. Carcinogenesis. 2000;21:497–503. - PubMed
-
- Kang Y, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. - PubMed
-
- Clark EA, Golub TR, Lander ES, Hynes RO. Genomic analysis of metastasis reveals an essential role for RhoC. Nature. 2000;406:532–535. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
