Global detection of molecular changes reveals concurrent alteration of several biological pathways in nonsmall cell lung cancer cells
- PMID: 16049682
- PMCID: PMC1544372
- DOI: 10.1007/s00438-005-0014-7
Global detection of molecular changes reveals concurrent alteration of several biological pathways in nonsmall cell lung cancer cells
Abstract
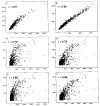
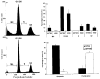
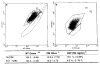
To identify the molecular changes that occur in non-small cell lung carcinoma (NSCLC), we compared the gene expression profile of the NCI-H292 (H292) NSCLC cell line with that of normal human tracheobronchial epithelial (NHTBE) cells. The NHTBE cells were grown in a three-dimensional organotypic culture system that permits maintenance of the normal pseudostratified mucociliary phenotype characteristic of bronchial epithelium in vivo. Microarray analysis using the Affymetrix oligonucleotide chip U95Av2 revealed that 1,683 genes showed a >1.5-fold change in expression in the H292 cell line relative to the NHTBE cells. Specifically, 418 genes were downregulated and 1,265 were upregulated in the H292 cells. The expression data for selected genes were validated in several different NSCLC cell lines using quantitative real-time PCR and Western analysis. Further analysis of the differentially expressed genes indicated that WNT responses, apoptosis, cell cycle regulation and cell proliferation were significantly altered in the H292 cells. Functional analysis using fluorescence-activated cell sorting confirmed concurrent changes in the activity of these pathways in the H292 line. These findings show that (1) NSCLC cells display deregulation of the WNT, apoptosis, proliferation and cell cycle pathways, as has been found in many other types of cancer cells, and (2) that organotypically cultured NHTBE cells can be used as a reference to identify genes and pathways that are differentially expressed in tumor cells derived from bronchogenic epithelium.
Figures



Similar articles
-
cDNA microarray analysis of the effect of cantharidin on DNA damage, cell cycle and apoptosis-associated gene expression in NCI-H460 human lung cancer cells in vitro.Mol Med Rep. 2015 Jul;12(1):1030-42. doi: 10.3892/mmr.2015.3538. Epub 2015 Mar 24. Mol Med Rep. 2015. PMID: 25815777 Free PMC article.
-
Expression of nicotinic acetylcholine receptor subunit genes in non-small-cell lung cancer reveals differences between smokers and nonsmokers.Cancer Res. 2007 May 15;67(10):4638-47. doi: 10.1158/0008-5472.CAN-06-4628. Cancer Res. 2007. PMID: 17510389
-
Diminished expression of S100A2, a putative tumor suppressor, at early stage of human lung carcinogenesis.Cancer Res. 2001 Nov 1;61(21):7999-8004. Cancer Res. 2001. PMID: 11691825
-
Mesenchymal stem cells in non-small cell lung cancer--different from others? Insights from comparative molecular and functional analyses.Lung Cancer. 2013 Apr;80(1):19-29. doi: 10.1016/j.lungcan.2012.12.015. Epub 2013 Jan 5. Lung Cancer. 2013. PMID: 23294501
-
Analysis of 20 genes at chromosome band 12q13: RACGAP1 and MCRS1 overexpression in nonsmall-cell lung cancer.Genes Chromosomes Cancer. 2013 Mar;52(3):305-15. doi: 10.1002/gcc.22030. Epub 2012 Dec 8. Genes Chromosomes Cancer. 2013. PMID: 23225332
Cited by
-
Human lung project: evaluating variance of gene expression in the human lung.Am J Respir Cell Mol Biol. 2006 Jul;35(1):65-71. doi: 10.1165/rcmb.2004-0261OC. Epub 2006 Feb 23. Am J Respir Cell Mol Biol. 2006. PMID: 16498083 Free PMC article.
-
Long noncoding RNA LINC00673-v4 promotes aggressiveness of lung adenocarcinoma via activating WNT/β-catenin signaling.Proc Natl Acad Sci U S A. 2019 Jul 9;116(28):14019-14028. doi: 10.1073/pnas.1900997116. Epub 2019 Jun 24. Proc Natl Acad Sci U S A. 2019. PMID: 31235588 Free PMC article.
-
Testicular germ cell tumor susceptibility genes from the consomic 129.MOLF-Chr19 mouse strain.Mamm Genome. 2007 Aug;18(8):584-95. doi: 10.1007/s00335-007-9036-2. Epub 2007 Aug 2. Mamm Genome. 2007. PMID: 17671812 Free PMC article.
-
Porphyromonas gingivalis gingipains potentially affect MUC5AC gene expression and protein levels in respiratory epithelial cells.FEBS Open Bio. 2021 Feb;11(2):446-455. doi: 10.1002/2211-5463.13066. Epub 2020 Dec 30. FEBS Open Bio. 2021. PMID: 33332733 Free PMC article.
-
Comparative study of gene set enrichment methods.BMC Bioinformatics. 2009 Sep 2;10:275. doi: 10.1186/1471-2105-10-275. BMC Bioinformatics. 2009. PMID: 19725948 Free PMC article.
References
-
- Amatschek S, Koenig U, Auer H, Steinlein P, Pacher M, Gruenfelder A, Dekan G, Vogl S, Kubista E, Heider KH, Stratowa C, Schreiber M, Sommergruber W. Tissue-wide expression profiling using cDNA subtraction and microarrays to identify tumor-specific genes. Cancer Res. 2004;64:844–856. - PubMed
-
- Beere HM, Wolf BB, Cain K, Mosser DD, Mahboubi A, Kuwana T, Tailor P, Morimoto RI, Cohen GM, Green DR. Heat-shock protein 70 inhibits apoptosis by preventing recruitment of procaspase-9 to the Apaf-1 apoptosome. Nat Cell Biol. 2000;2:469–475. - PubMed
-
- Braakhuis BJ, Tabor MP, Kummer JA, Leemans CR, Brakenhoff RH. A genetic explanation of Slaughter’s concept of field cancerization: evidence and clinical implications. Cancer Res. 2003;63:1727–1730. - PubMed
-
- Braakhuis BJ, Leemans CR, Brakenhoff RH. Using tissue adjacent to carcinoma as a normal control: an obvious but questionable practice. J Pathol. 2004;203:620–621. - PubMed
-
- Broers JL, Viallet J, Jensen SM, Pass H, Travis WD, Minna JD, Linnoila RI. Expression of c-myc in progenitor cells of the bronchopulmonary epithelium and in a large number of non-small cell lung cancers. Am J Respir Cell Mol Biol. 1993;9:33–43. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

