Mutagenesis of the murine hepatitis virus nsp1-coding region identifies residues important for protein processing, viral RNA synthesis, and viral replication
- PMID: 16051301
- PMCID: PMC7111834
- DOI: 10.1016/j.virol.2005.06.035
Mutagenesis of the murine hepatitis virus nsp1-coding region identifies residues important for protein processing, viral RNA synthesis, and viral replication
Abstract
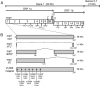
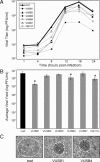
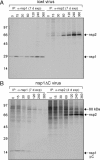
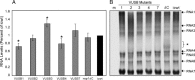
Despite ongoing research investigating mechanisms of coronavirus replication, functions of many viral nonstructural proteins (nsps) remain unknown. In the current study, a reverse genetic approach was used to define the role of the 28-kDa amino-terminal product (nsp1) of the gene 1 polyprotein during replication of the coronavirus murine hepatitis virus (MHV) in cell culture. To determine whether nsp1 is required for MHV replication and to identify residues critical for protein function, mutant viruses that contained deletions or point mutations within the nsp1-coding region were generated and assayed for defects in viral replication, viral protein expression, protein localization, and RNA synthesis. The results demonstrated that the carboxy-terminal half of nsp1 (residues K(124) through L(241)) was dispensable for virus replication in culture but was required for efficient proteolytic cleavage of nsp1 from the gene 1 polyprotein and for optimal viral replication. Furthermore, whereas deletion of nsp1 residues amino-terminal to K(124) failed to produce infectious virus, point mutagenesis of the nsp1 amino-terminus allowed recovery of several mutants with altered replication and RNA synthesis. This study identifies nsp1 residues important for protein processing, viral RNA synthesis, and viral replication.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

