Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies
- PMID: 16051804
- PMCID: PMC1182643
- DOI: 10.1128/JVI.79.16.10108-10125.2005
Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies
Abstract
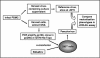
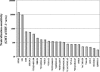
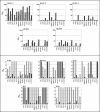
Induction of broadly cross-reactive neutralizing antibodies is a high priority for AIDS vaccine development but one that has proven difficult to be achieved. While most immunogens generate antibodies that neutralize a subset of T-cell-line-adapted strains of human immunodeficiency virus type 1 (HIV-1), none so far have generated a potent, broadly cross-reactive response against primary isolates of the virus. Even small increments in immunogen improvement leading to increases in neutralizing antibody titers and cross-neutralizing activity would accelerate vaccine development; however, a lack of uniformity in target strains used by different investigators to assess cross-neutralization has made the comparison of vaccine-induced antibody responses difficult. Thus, there is an urgent need to establish standard panels of HIV-1 reference strains for wide distribution. To facilitate this, full-length gp160 genes were cloned from acute and early subtype B infections and characterized for use as reference reagents to assess neutralizing antibodies against clade B HIV-1. Individual gp160 clones were screened for infectivity as Env-pseudotyped viruses in a luciferase reporter gene assay in JC53-BL (TZM-bl) cells. Functional env clones were sequenced and their neutralization phenotypes characterized by using soluble CD4, monoclonal antibodies, and serum samples from infected individuals and noninfected recipients of a recombinant gp120 vaccine. Env clones from 12 R5 primary HIV-1 isolates were selected that were not unusually sensitive or resistant to neutralization and comprised a wide spectrum of genetic, antigenic, and geographic diversity. These reference reagents will facilitate proficiency testing and other validation efforts aimed at improving assay performance across laboratories and can be used for standardized assessments of vaccine-elicited neutralizing antibodies.
Figures




References
-
- Arthur, L. O., J. W. Bess, Jr., E. N. Chertova, J. L. Rossio, M. T. Esser, R. E. Benveniste, L. E. Henderson, and J. D. Lifson. 1998. Chemical inactivation of retroviral infectivity by targeting nucleocapsid protein zinc fingers: a candidate SIV vaccine. AIDS Res. Hum. Retroviruses 14(Suppl. 3):S311-S319. - PubMed
-
- Back, N. K. T., L. Smit, J.-J. de Jong, W. Keulen, M. Schutten, J. Goudsmit, and M. Tersmette. 1994. An N-glycan within the human immunodeficiency virus type 1 gp120 V3 loop affects virus neutralization. Virology 199:431-438. - PubMed
-
- Barbato, G., E. Bianchi, P. Ingallinella, W. H. Hurn, M. D. Miller, G. Ciliberto, R. Cortese, R. Bazzo, J. W. Shiver, and A. Pessi. 2003. Structural analysis of the epitope of the anti-HIV antibody 2F5 sheds light into its mechanism of neutralization and HIV fusion. J. Mol. Biol. 330:1101-1115. - PubMed
-
- Barnett, S. W., S. Lu, I. Sravastava, S. Cherelis, A. Gettie, J. Blanchard, S. Wang, I. Mboudjeka, L. Leung, Y. Lian, A. Fong, C. Buckner, A. Ly, S. Hilt, J. Ulmer, C. T. Wild, J. R. Mascola, and L. Stamatatos. 2001. The ability of an oligomeric human immunodeficiency virus type 1 (HIV-1) envelope antigen to elicit neutralizing antibodies against primary HIV-1 isolates is improved following partial deletion of the second hypervariable region. J. Virol. 75:5526-5540. - PMC - PubMed
-
- Beddows, S., S. Lister, R. Cheingsong, C. Bruck, and J. Weber. 1999. Comparison of the antibody repertoire generated in healthy volunteers following immunization with a monomeric recombinant gp120 construct derived from a CCR5/CXCR4-using human immunodeficiency virus type 1 isolate with sera from naturally infected individuals. J. Virol. 73:1740-1745. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 AI027767/AI/NIAID NIH HHS/United States
- AI85338/AI/NIAID NIH HHS/United States
- AI46705/AI/NIAID NIH HHS/United States
- AI41530/AI/NIAID NIH HHS/United States
- AI055386/AI/NIAID NIH HHS/United States
- N01 AI085338/AI/NIAID NIH HHS/United States
- R01 AI047708/AI/NIAID NIH HHS/United States
- R56 AI047708/AI/NIAID NIH HHS/United States
- U01 AI041530/AI/NIAID NIH HHS/United States
- R21 AI055386/AI/NIAID NIH HHS/United States
- AI54497/AI/NIAID NIH HHS/United States
- N01 AI030034/AI/NIAID NIH HHS/United States
- AI47708/AI/NIAID NIH HHS/United States
- AI27767/AI/NIAID NIH HHS/United States
- AI27742/AI/NIAID NIH HHS/United States
- P30 AI027742/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

