Evidence for frequent reinfection with human immunodeficiency virus type 1 of a different subtype
- PMID: 16051862
- PMCID: PMC1182664
- DOI: 10.1128/JVI.79.16.10701-10708.2005
Evidence for frequent reinfection with human immunodeficiency virus type 1 of a different subtype
Abstract
A major premise underlying current human immunodeficiency virus type 1 (HIV-1) vaccine approaches is that preexisting HIV-1-specific immunity will block or reduce infection. However, the recent identification of several cases of HIV-1 reinfection suggests that the specific immune response generated for chronic HIV-1 infection may not be adequate to protect against infection by a second HIV-1 strain. It has been unclear, though, whether these individuals are representative of the global epidemic or are rare cases. Here we show that in a population of high-risk women, HIV-1 reinfection occurs almost as commonly as first infections. The study was designed to detect cases of reinfection by HIV-1 of a different subtype and thus captured cases where there was considerable diversity between the first and second strain. In each case, the second virus emerged approximately 1 year after the first infection, and in two cases, it emerged when viral levels were high, suggesting that a well-established HIV-1 infection may provide little benefit in terms of immunizing against reinfection, at least by more-divergent HIV-1 variants. Our findings indicate an urgent need for studies of larger cohorts to determine the incidence and timing of both intersubtype and intrasubtype reinfection.
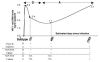
Figures




References
-
- Altfeld, M., T. M. Allen, X. G. Yu, M. N. Johnston, D. Agrawal, B. T. Korber, D. C. Montefiori, D. H. O'Connor, B. T. Davis, P. K. Lee, E. L. Maier, J. Harlow, P. J. Goulder, C. Brander, E. S. Rosenberg, and B. D. Walker. 2002. HIV-1 superinfection despite broad CD8+ T-cell responses containing replication of the primary virus. Nature 420:434-439. - PubMed
-
- Artenstein, A. W., T. C. VanCott, J. R. Mascola, J. K. Carr, P. A. Hegerich, J. Gaywee, E. Sanders-Buell, M. L. Robb, D. E. Dayhoff, S. Thitivichianlert, et al. 1995. Dual infection with human immunodeficiency virus type 1 of distinct envelope subtypes in humans. J. Infect. Dis. 171:805-810. - PubMed
-
- Baeten, J. M., and J. Overbaugh. 2003. Measuring the infectiousness of persons with HIV-1: opportunities for preventing sexual HIV-1 transmission. Curr. HIV Res. 1:69-86. - PubMed
-
- Butto, S., C. Argentini, A. M. Mazzella, M. P. Iannotti, P. Leone, A. Nicolosi, and G. Rezza. 1997. Dual infection with different strains of the same HIV-1 subtype. AIDS 11:694-696. - PubMed
-
- Chakraborty, B., L. Valer, C. De Mendoza, V. Soriano, and M. E. Quinones-Mateu. 2004. Failure to detect human immunodeficiency virus type 1 superinfection in 28 HIV-seroconcordant individuals with high risk of reexposure to the virus. AIDS Res. Hum. Retrovir. 20:1026-1031. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

