Mechanisms underlying the transcriptional regulation of skeletal myogenesis
- PMID: 16055324
- PMCID: PMC1283108
- DOI: 10.1016/j.gde.2005.04.015
Mechanisms underlying the transcriptional regulation of skeletal myogenesis
Abstract
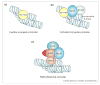
During skeletal myogenesis, chromatin-modifying enzymes are engaged at discrete genomic regions by transcription factors that recognize sequence-specific DNA motifs located at muscle gene regulatory regions. The composition of the chromatin-bound protein complexes and their temporally and spatially regulated recruitment influence gene expression. Recent findings are consistent with the concept that chromatin modifiers play an important role in regulating skeletal muscle gene expression and cellular differentiation.
Figures


References
-
- Gill G, Something about SUMO inhibits transcription Curr Opin Genet Dev 2005, 15: in press. - PubMed
-
- Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J Biol Chem. 2002;277:45099–45107. - PubMed
-
- Fulco M, Schiltz RL, Iezzi S, King MT, Zhao P, Kashiwaya Y, Hoffman E, Veech RL, Sartorelli V. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol Cell. 2003;12:51–62. - PubMed
-
- Thompson PR, Wang D, Wang L, Fulco M, Pediconi N, Zhang D, An W, Ge Q, Roeder RG, Wong J, et al. Regulation of the p300 HAT domain via a novel activation loop. Nat Struct Mol Biol. 2004;11:308–315. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

