Nuclear aggresomes form by fusion of PML-associated aggregates
- PMID: 16055507
- PMCID: PMC1237092
- DOI: 10.1091/mbc.e05-01-0019
Nuclear aggresomes form by fusion of PML-associated aggregates
Abstract
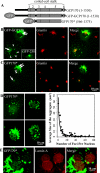
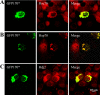
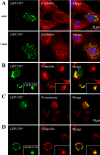
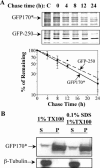
Nuclear aggregates formed by proteins containing expanded poly-glutamine (poly-Q) tracts have been linked to the pathogenesis of poly-Q neurodegenerative diseases. Here, we show that a protein (GFP170*) lacking poly-Q tracts forms nuclear aggregates that share characteristics of poly-Q aggregates. GFP170* aggregates recruit cellular chaperones and proteasomes, and alter the organization of nuclear domains containing the promyelocytic leukemia (PML) protein. These results suggest that the formation of nuclear aggregates and their effects on nuclear architecture are not specific to poly-Q proteins. Using GFP170* as a model substrate, we explored the mechanistic details of nuclear aggregate formation. Fluorescence recovery after photobleaching and fluorescence loss in photobleaching analyses show that GFP170* molecules exchange rapidly between aggregates and a soluble pool of GFP170*, indicating that the aggregates are dynamic accumulations of GFP170*. The formation of cytoplasmic and nuclear GFP170* aggregates is microtubule-dependent. We show that within the nucleus, GFP170* initially deposits in small aggregates at or adjacent to PML bodies. Time-lapse imaging of live cells shows that small aggregates move toward each other and fuse to form larger aggregates. The coalescence of the aggregates is accompanied by spatial rearrangements of the PML bodies. Significantly, we find that the larger nuclear aggregates have complex internal substructures that reposition extensively during fusion of the aggregates. These studies suggest that nuclear aggregates may be viewed as dynamic multidomain inclusions that continuously remodel their components.
Figures



References
-
- Bence, N. F., Sampat, R. M., and Kopito, R. R. (2001). Impairment of the ubiquitin-proteasome system by protein aggregation. Science 292, 1552-1555. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

