The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway
- PMID: 16055633
- PMCID: PMC1197433
- DOI: 10.1105/tpc.105.034009
The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway
Abstract
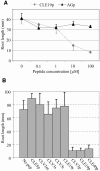
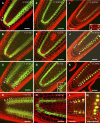
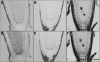
CLAVATA3 (CLV3), CLV3/ESR19 (CLE19), and CLE40 belong to a family of 26 genes in Arabidopsis thaliana that encode putative peptide ligands with unknown identity. It has been shown previously that ectopic expression of any of these three genes leads to a consumption of the root meristem. Here, we show that in vitro application of synthetic 14-amino acid peptides, CLV3p, CLE19p, and CLE40p, corresponding to the conserved CLE motif, mimics the overexpression phenotype. The same result was observed when CLE19 protein was applied externally. Interestingly, clv2 failed to respond to the peptide treatment, suggesting that CLV2 is involved in the CLE peptide signaling. Crossing of the CLE19 overexpression line with clv mutants confirms the involvement of CLV2. Analyses using tissue-specific marker lines revealed that the peptide treatments led to a premature differentiation of the ground tissue daughter cells and misspecification of cell identity in the pericycle and endodermis layers. We propose that these 14-amino acid peptides represent the major active domain of the corresponding CLE proteins, which interact with or saturate an unknown cell identity-maintaining CLV2 receptor complex in roots, leading to consumption of the root meristem.
Figures




References
-
- Alberts, B., Bray, D., Lewis, J., Raff, M., Roberts, K., and Watson, J.D. (1994). Molecular Biology of the Cell. (New York: Garland).
-
- Birnbaum, K., Shasha, D.E., Wang, J.Y., Jung, J.W., Lambert, G.M., Galbraith, D.W., and Benfey, P.N. (2003). A gene expression map of the Arabidopsis root. Science 302, 1956–1960. - PubMed
-
- Bonello, J.F., Sevilla-Lecoq, S., Berne, A., Risueno, M.C., Dumas, C., and Rogowsky, P.M. (2002). Esr proteins are secreted by the cells of the embryo surrounding region. J. Exp. Bot. 53, 1559–1568. - PubMed
-
- Brand, U., Fletcher, J.C., Hobe, M., Meyerowitz, E.M., and Simon, R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289, 617–619. - PubMed
-
- Casamitjana-Martinez, E., Hofhuis, H.F., Xu, J., Liu, C.M., Heidstra, R., and Scheres, B. (2003). Root-specific CLE19 over-expression and the sol1 and sol2 suppressors implicate a CLV-like pathway in the control of Arabidopsis root meristem maintenances. Curr. Biol. 13, 1435–1441. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

