An atypical RNA pseudoknot stimulator and an upstream attenuation signal for -1 ribosomal frameshifting of SARS coronavirus
- PMID: 16055920
- PMCID: PMC1182165
- DOI: 10.1093/nar/gki731
An atypical RNA pseudoknot stimulator and an upstream attenuation signal for -1 ribosomal frameshifting of SARS coronavirus
Abstract
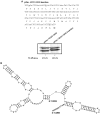
The -1 ribosomal frameshifting requires the existence of an in cis RNA slippery sequence and is promoted by a downstream stimulator RNA. An atypical RNA pseudoknot with an extra stem formed by complementary sequences within loop 2 of an H-type pseudoknot is characterized in the severe acute respiratory syndrome coronavirus (SARS CoV) genome. This pseudoknot can serve as an efficient stimulator for -1 frameshifting in vitro. Mutational analysis of the extra stem suggests frameshift efficiency can be modulated via manipulation of the secondary structure within the loop 2 of an infectious bronchitis virus-type pseudoknot. More importantly, an upstream RNA sequence separated by a linker 5' to the slippery site is also identified to be capable of modulating the -1 frameshift efficiency. RNA sequence containing this attenuation element can downregulate -1 frameshifting promoted by an atypical pseudoknot of SARS CoV and two other pseudoknot stimulators. Furthermore, frameshift efficiency can be reduced to half in the presence of the attenuation signal in vivo. Therefore, this in cis RNA attenuator represents a novel negative determinant of general importance for the regulation of -1 frameshift efficiency, and is thus a potential antiviral target.
Figures





References
-
- Gesteland R.F., Atkins J.F. Recoding: dynamic reprogramming of translation. Annu. Rev. Biochem. 1996;65:741–768. - PubMed
-
- Harger J.W., Meskauskas A., Dinman J.D. An ‘integrated model’ of programmed ribosomal frameshifting. Trends Biochem. Sci. 2002;27:448–454. - PubMed
-
- Namy O., Rousset J.P., Napthine S., Brierley I. Reprogrammed genetic decoding in cellular gene expression. Mol. Cell. 2004;13:157–168. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

