Weighing in on ubiquitin: the expanding role of mass-spectrometry-based proteomics
- PMID: 16056266
- PMCID: PMC1224607
- DOI: 10.1038/ncb0805-750
Weighing in on ubiquitin: the expanding role of mass-spectrometry-based proteomics
Abstract
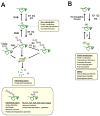
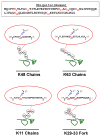
Mass-spectrometry-based proteomics has become an essential tool for the qualitative and quantitative analysis of cellular systems. The biochemical complexity and functional diversity of the ubiquitin system are well suited to proteomic studies. This review summarizes advances involving the identification of ubiquitinated proteins, the elucidation of ubiquitin-modification sites and the determination of polyubiquitin chain linkages, as well as offering a perspective on the application of emerging technologies for mechanistic and functional studies of protein ubiquitination.
Figures


References
-
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. - PubMed
-
- Pickart CM. Mechanisms underlying Ubiquitination. Nat Rev Cell Mol Biol. 2001;70:503–533. - PubMed
-
- Finley D, Ciechanover A, Varshavsky A. Ubiquitin as a central cellular regulator. Cell. 2004;116:S29–32. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

