Parallel beta-sheets and polar zippers in amyloid fibrils formed by residues 10-39 of the yeast prion protein Ure2p
- PMID: 16060675
- PMCID: PMC1380259
- DOI: 10.1021/bi050724t
Parallel beta-sheets and polar zippers in amyloid fibrils formed by residues 10-39 of the yeast prion protein Ure2p
Abstract
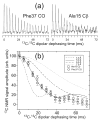
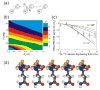
We report the results of solid-state nuclear magnetic resonance (NMR) and atomic force microscopy measurements on amyloid fibrils formed by residues 10-39 of the yeast prion protein Ure2p (Ure2p(10)(-)(39)). Measurements of intermolecular (13)C-(13)C nuclear magnetic dipole-dipole couplings indicate that Ure2p(10)(-)(39) fibrils contain in-register parallel beta-sheets. Measurements of intermolecular (15)N-(13)C dipole-dipole couplings, using a new solid-state NMR technique called DSQ-REDOR, are consistent with hydrogen bonds between side chain amide groups of Gln18 residues. Such side chain hydrogen bonding interactions have been called "polar zippers" by M. F. Perutz and have been proposed to stabilize amyloid fibrils formed by peptides with glutamine- and asparagine-rich sequences, such as Ure2p(10)(-)(39). We propose that polar zipper interactions account for the in-register parallel beta-sheet structure in Ure2p(10)(-)(39) fibrils and that similar peptides will also exhibit parallel beta-sheet structures in amyloid fibrils. We present molecular models for Ure2p(10)(-)(39) fibrils that are consistent with available experimental data. Finally, we show that solid-state (13)C NMR chemical shifts for (13)C-labeled Ure2p(10)(-)(39) fibrils are insensitive to hydration level, indicating that the fibril structure is not affected by the presence or absence of bulk water.
Figures






Similar articles
-
Characterization of beta-sheet structure in Ure2p1-89 yeast prion fibrils by solid-state nuclear magnetic resonance.Biochemistry. 2007 Nov 13;46(45):13149-62. doi: 10.1021/bi700826b. Epub 2007 Oct 23. Biochemistry. 2007. PMID: 17953455
-
The core of Ure2p prion fibrils is formed by the N-terminal segment in a parallel cross-β structure: evidence from solid-state NMR.J Mol Biol. 2011 Jun 3;409(2):263-77. doi: 10.1016/j.jmb.2011.03.067. Epub 2011 Apr 8. J Mol Biol. 2011. PMID: 21497604 Free PMC article.
-
Amyloid of the prion domain of Sup35p has an in-register parallel beta-sheet structure.Proc Natl Acad Sci U S A. 2006 Dec 26;103(52):19754-9. doi: 10.1073/pnas.0609638103. Epub 2006 Dec 14. Proc Natl Acad Sci U S A. 2006. PMID: 17170131 Free PMC article.
-
Molecular structures of amyloid and prion fibrils: consensus versus controversy.Acc Chem Res. 2013 Jul 16;46(7):1487-96. doi: 10.1021/ar300282r. Epub 2013 Jan 7. Acc Chem Res. 2013. PMID: 23294335 Free PMC article. Review.
-
Molecular structure of amyloid fibrils: insights from solid-state NMR.Q Rev Biophys. 2006 Feb;39(1):1-55. doi: 10.1017/S0033583506004173. Epub 2006 Jun 13. Q Rev Biophys. 2006. PMID: 16772049 Review.
Cited by
-
Primary sequence independence for prion formation.Proc Natl Acad Sci U S A. 2005 Sep 6;102(36):12825-30. doi: 10.1073/pnas.0506136102. Epub 2005 Aug 25. Proc Natl Acad Sci U S A. 2005. PMID: 16123127 Free PMC article.
-
Molecular alignment within beta-sheets in Abeta(14-23) fibrils: solid-state NMR experiments and theoretical predictions.Biophys J. 2007 Jan 15;92(2):594-602. doi: 10.1529/biophysj.106.091017. Epub 2006 Oct 20. Biophys J. 2007. PMID: 17056725 Free PMC article.
-
Repeat domains of melanosome matrix protein Pmel17 orthologs form amyloid fibrils at the acidic melanosomal pH.J Biol Chem. 2011 Mar 11;286(10):8385-8393. doi: 10.1074/jbc.M110.197152. Epub 2010 Dec 10. J Biol Chem. 2011. PMID: 21148556 Free PMC article.
-
Anti-Prion Systems in Saccharomyces cerevisiae.J Neurochem. 2025 Mar;169(3):e70045. doi: 10.1111/jnc.70045. J Neurochem. 2025. PMID: 40130511 Free PMC article. Review.
-
High-resolution MAS NMR analysis of PI3-SH3 amyloid fibrils: backbone conformation and implications for protofilament assembly and structure.Biochemistry. 2010 Sep 7;49(35):7474-84. doi: 10.1021/bi100864t. Biochemistry. 2010. PMID: 20707313 Free PMC article.
References
-
- Lansbury PT, Costa PR, Griffiths JM, Simon EJ, Auger M, Halverson KJ, Kocisko DA, Hendsch ZS, Ashburn TT, Spencer RGS, Tidor B, Griffin RG. Structural model for the β-amyloid fibril based on interstrand alignment of an antiparallel-sheet comprising a C-terminal peptide. Nat Struct Biol. 1995;2:990–998. - PubMed
-
- Sunde M, Serpell LC, Bartlam M, Fraser PE, Pepys MB, Blake CCF. Common core structure of amyloid fibrils by synchrotron x-ray diffraction. J Mol Biol. 1997;273:729–739. - PubMed
-
- Gregory DM, Benzinger TLS, Burkoth TS, Miller-Auer H, Lynn DG, Meredith SC, Botto RE. Dipolar recoupling NMR of biomolecular self-assemblies: Determining inter- and intrastrand distances in fibrilized Alzheimer’s β-amyloid peptide. Solid State Nucl Magn Reson. 1998;13:149–166. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

