RNase P cleaves transient structures in some riboswitches
- PMID: 16061811
- PMCID: PMC1183601
- DOI: 10.1073/pnas.0505271102
RNase P cleaves transient structures in some riboswitches
Abstract
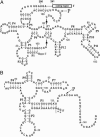
RNase P from Escherichia coli cleaves the coenzyme B12 riboswitch from E. coli and a similar one from Bacillus subtilis. The cleavage sites do not occur in any recognizable structure, as judged from theoretical schemes that have been drawn for these 5' UTRs. However, it is possible to draw a scheme that is a good representation of the E. coli cleavage site for RNase P and for the cleavage site in B. subtilis. These data indicate that transient structures are important in RNase P cleavage and in riboswitch function. Coenzyme B12 has a small inhibitory effect on E. coli RNase P cleavage of the E. coli riboswitch. Both E. coli RNase P and a partially purified RNase P from Aspergillus nidulans mycelia succeeded in cleaving a putative arginine riboswitch from A. nidulans. The cleavage site may be a representative of another model substrate for eukaryotic RNase P. This 5' UTR controls splicing of the arginase mRNA in A. nidulans. Four other riboswitches in E. coli were not cleaved by RNase P under the conditions tested.
Figures



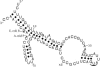
References
-
- Winkler, W. C. & Breaker, R. R. (2003) ChemBioChem 4, 1024–1032. - PubMed
-
- Altman, S. & Kirsebom, L. (1999) in The RNA World, eds. Gesteland, R., Cech, T. & Atkins, J. F. (Cold Spring Harbor Lab. Press, Woodbury, NY), 2nd Ed., pp. 351–380.
-
- Borsuk, P., Dzikowska, A., Empel, J., Grzelak, A., Grzeskowiak, R. & Weglenski, P. (1999) Acta Biochim. Polon. 46, 391–403. - PubMed
-
- Li, Y. & Altman, S. (2004) J. Mol. Biol. 339, 31–39. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

