Drosophila host defense after oral infection by an entomopathogenic Pseudomonas species
- PMID: 16061818
- PMCID: PMC1183552
- DOI: 10.1073/pnas.0502240102
Drosophila host defense after oral infection by an entomopathogenic Pseudomonas species
Abstract
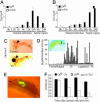
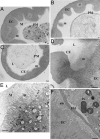
Drosophila has been shown to be a valuable model for the investigation of host-pathogen interactions. Study of the Drosophila immune response has been hampered, however, by the lack of true Drosophila pathogens. In nearly all studies reported, the bacteria used were directly injected within the body cavity of the insect, bypassing the initial steps of a natural interaction. Here, we report the identification of a previously uncharacterized bacterial species, Pseudomonas entomophila (Pe), which has the capacity to induce the systemic expression of antimicrobial peptide genes in Drosophila after ingestion. In contrast to previously identified bacteria, Pe is highly pathogenic to both Drosophila larvae and adults, and its persistence in larvae leads to a massive destruction of gut cells. Using this strain, we have analyzed the modulation of the larval transcriptome upon bacterial infection. We found that natural infection by Pe induces a dramatic change in larval gene expression. In addition to immunity genes, our study identifies many genes associated with Pe pathogenesis that have been previously unreported.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

