NuMA is a major acceptor of poly(ADP-ribosyl)ation by tankyrase 1 in mitosis
- PMID: 16076287
- PMCID: PMC1276914
- DOI: 10.1042/BJ20050885
NuMA is a major acceptor of poly(ADP-ribosyl)ation by tankyrase 1 in mitosis
Abstract
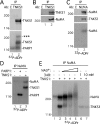
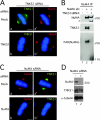
Tankyrase 1 is a PARP [poly(ADP-ribose) polymerase] that localizes to multiple subcellular sites, including telomeres and mitotic centrosomes. Previous studies demonstrated that cells deficient in tankyrase 1 suffered a block in resolution of sister telomeres and arrested in early anaphase [Dynek and Smith (2004) Science 304, 97-100]. This phenotype was dependent on the catalytic PARP activity of tankyrase 1. To identify critical acceptors of PARsylation [poly(ADP-ribosyl)ation] by tankyrase 1 in mitosis, tankyrase 1 immunoprecipitates were analysed for associated PARsylated proteins. We identified NuMA (nuclear mitotic apparatus protein) as a major acceptor of poly(ADP-ribose) from tankyrase 1 in mitosis. We showed by immunofluorescence and immunoprecipitation that association between tankyrase 1 and NuMA increases dramatically at the onset of mitosis, concomitant with PARsylation of NuMA. Knockdown of tankyrase 1 by siRNA (small interfering RNA) eliminates PARsylation of NuMA in mitosis, confirming tankyrase 1 as the PARP responsible for this modification. However, even in the absence of tankyrase 1 and PARsylation, NuMA localizes to spindle poles. By contrast, siRNA knockdown of NuMA results in complete loss of tankyrase 1 from spindle poles. We discuss our result in terms of a model where PARsylation of NuMA by tankyrase 1 in mitosis could play a role in sister telomere separation and/or mitotic progression.
Figures

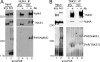

Comment in
-
Regulation of mitosis by poly(ADP-ribosyl)ation.Biochem J. 2005 Oct 15;391(Pt 2):e5-6. doi: 10.1042/BJ20051437. Biochem J. 2005. PMID: 16212557 Free PMC article.
Similar articles
-
Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function.Nat Cell Biol. 2005 Nov;7(11):1133-9. doi: 10.1038/ncb1322. Nat Cell Biol. 2005. PMID: 16244666
-
Regulation of mitosis by poly(ADP-ribosyl)ation.Biochem J. 2005 Oct 15;391(Pt 2):e5-6. doi: 10.1042/BJ20051437. Biochem J. 2005. PMID: 16212557 Free PMC article.
-
GDP-mannose-4,6-dehydratase is a cytosolic partner of tankyrase 1 that inhibits its poly(ADP-ribose) polymerase activity.Mol Cell Biol. 2012 Aug;32(15):3044-53. doi: 10.1128/MCB.00258-12. Epub 2012 May 29. Mol Cell Biol. 2012. PMID: 22645305 Free PMC article.
-
Tankyrase function at telomeres, spindle poles, and beyond.Biochimie. 2008 Jan;90(1):83-92. doi: 10.1016/j.biochi.2007.07.012. Epub 2007 Jul 24. Biochimie. 2008. PMID: 17825467 Review.
-
Tankyrases as drug targets.FEBS J. 2013 Aug;280(15):3576-93. doi: 10.1111/febs.12320. Epub 2013 Jun 18. FEBS J. 2013. PMID: 23648170 Review.
Cited by
-
The DNA damage-inducible C. elegans tankyrase is a nuclear protein closely linked to chromosomes.Mol Cell Biochem. 2009 Apr;324(1-2):73-83. doi: 10.1007/s11010-008-9986-z. Epub 2008 Dec 23. Mol Cell Biochem. 2009. PMID: 19104912
-
Tankyrase-1 function at telomeres and during mitosis is regulated by Polo-like kinase-1-mediated phosphorylation.Cell Death Differ. 2012 Feb;19(2):321-32. doi: 10.1038/cdd.2011.101. Epub 2011 Aug 5. Cell Death Differ. 2012. PMID: 21818122 Free PMC article.
-
Protein requirements for sister telomere association in human cells.EMBO J. 2007 Nov 28;26(23):4867-78. doi: 10.1038/sj.emboj.7601903. Epub 2007 Oct 25. EMBO J. 2007. PMID: 17962804 Free PMC article.
-
Poly(ADP-ribose) polymerase enzymes and the maintenance of genome integrity.Cell Mol Life Sci. 2020 Jan;77(1):19-33. doi: 10.1007/s00018-019-03366-0. Epub 2019 Nov 21. Cell Mol Life Sci. 2020. PMID: 31754726 Free PMC article. Review.
-
The phenanthrene derivative PJ34 exclusively eradicates human pancreatic cancer cells in xenografts.Oncotarget. 2019 Oct 22;10(58):6269-6282. doi: 10.18632/oncotarget.27268. eCollection 2019 Oct 22. Oncotarget. 2019. PMID: 31692907 Free PMC article.
References
-
- Smith S., Giriat I., Schmitt A., de Lange T. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science. 1998;282:1484–1487. - PubMed
-
- Smith S., de Lange T. Tankyrase promotes telomere elongation in human cells. Curr. Biol. 2000;10:1299–1302. - PubMed
-
- Dynek J. N., Smith S. Resolution of sister telomere association is required for progression through mitosis. Science. 2004;304:97–100. - PubMed
-
- de Lange T. Protection of mammalian telomeres. Oncogene. 2002;21:532–540. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

