The agr radiation: an early event in the evolution of staphylococci
- PMID: 16077103
- PMCID: PMC1196086
- DOI: 10.1128/JB.187.16.5585-5594.2005
The agr radiation: an early event in the evolution of staphylococci
Abstract
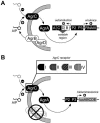
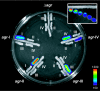
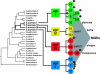
Agr is a global regulatory system in the staphylococci, operating by a classical two-component signaling module and controlling the expression of most of the genes encoding extracellular virulence factors. As it is autoinduced by a peptide, encoded within the locus, that is the ligand for the signal receptor, it is a sensor of population density or a quorum sensor and is the only known quorum-sensing system in the genus. agr is conserved throughout the staphylococci but has diverged along lines that appear to parallel speciation and subspeciation within the genus. This divergence has given rise to a novel type of interstrain and interspecies cross-inhibition that represents a fundamental aspect of the organism's biology and may be a predominant feature of the evolutionary forces that have driven it. We present evidence, using a newly developed, luciferase-based agr typing scheme, that the evolutionary divergence of the agr system was an early event in the evolution of the staphylococci and long preceded the development of the nucleotide polymorphisms presently used for genotyping. These polymorphisms developed, for the most part, within different agr groups; mobile genetic elements appear also to have diffused recently and, with a few notable exceptions, have come to reside largely indiscriminately within the several agr groups.
Figures
References
-
- Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. - PubMed
-
- Chu, M. C., M. E. Melish, and J. F. James. 1985. Tryprophan auxotypy associated with Staphylococcus aureus that produce toxic shock syndrome toxin. J. Infect. Dis. 151:1157-1158. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

