Coordinated replication and sequestration of oriC and dnaA are required for maintaining controlled once-per-cell-cycle initiation in Escherichia coli
- PMID: 16077105
- PMCID: PMC1196069
- DOI: 10.1128/JB.187.16.5605-5613.2005
Coordinated replication and sequestration of oriC and dnaA are required for maintaining controlled once-per-cell-cycle initiation in Escherichia coli
Abstract
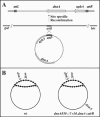
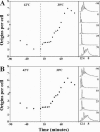
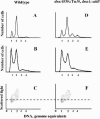
Escherichia coli cells were constructed in which the dnaA gene was moved to a location opposite oriC on the circular chromosome. In these cells the dnaA gene was replicated with significant delay relative to the origin. Consequently, the period where the newly replicated and hemimethylated oriC was sequestered no longer coincided with the period where the dnaA gene promoter was sequestered. DnaA protein synthesis was therefore expected to continue during origin sequestration. Despite a normal length of the sequestration period in such cells, they had increased origin content and also displayed asynchrony of initiation. This indicated that reinitiation occasionally occurred at some origins within the same cell cycle. The extra initiations took place in spite of a reduction in total DnaA protein concentration to about half of the wild-type level. We propose that this more efficient utilization of DnaA protein results from an increased availability at the end of the origin sequestration period. Therefore, coordinated sequestration of oriC and dnaA is required for maintaining controlled once-per-cell-cycle initiation.
Figures
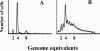


Similar articles
-
An oriC-like distribution of GATC sites mediates full sequestration of non-origin sequences in Escherichia coli.J Mol Biol. 2005 Jul 1;350(1):7-11. doi: 10.1016/j.jmb.2005.04.055. J Mol Biol. 2005. PMID: 15922360
-
The initiator protein DnaA contributes to keeping new origins inactivated by promoting the presence of hemimethylated DNA.J Mol Biol. 2008 Dec 31;384(5):1076-85. doi: 10.1016/j.jmb.2008.09.042. Epub 2008 Sep 25. J Mol Biol. 2008. PMID: 18835566
-
Role of DNA methylation at GATC sites in the dnaA promoter, dnaAp2.J Mol Microbiol Biotechnol. 2000 Jul;2(3):301-10. J Mol Microbiol Biotechnol. 2000. PMID: 10937439
-
Regulation of the initiation of chromosomal replication in bacteria.FEMS Microbiol Rev. 2007 Jul;31(4):378-87. doi: 10.1111/j.1574-6976.2007.00070.x. Epub 2007 Apr 25. FEMS Microbiol Rev. 2007. PMID: 17459114 Review.
-
[Initiation of DNA replication in Streptomyces; organizational comparison of the oriC region in prokaryotic organisms].Postepy Hig Med Dosw. 1995;49(1):149-60. Postepy Hig Med Dosw. 1995. PMID: 8657612 Review. Polish.
Cited by
-
Once in a lifetime: strategies for preventing re-replication in prokaryotic and eukaryotic cells.EMBO Rep. 2008 Feb;9(2):151-6. doi: 10.1038/sj.embor.2008.2. EMBO Rep. 2008. PMID: 18246107 Free PMC article. Review.
-
Evidence for roles of the Escherichia coli Hda protein beyond regulatory inactivation of DnaA.Mol Microbiol. 2012 Aug;85(4):648-68. doi: 10.1111/j.1365-2958.2012.08129.x. Epub 2012 Jul 13. Mol Microbiol. 2012. PMID: 22716942 Free PMC article.
-
Synchronous replication initiation in novel Mycobacterium tuberculosis dnaA cold-sensitive mutants.Mol Microbiol. 2009 Jan;71(2):291-304. doi: 10.1111/j.1365-2958.2008.06523.x. Epub 2008 Nov 14. Mol Microbiol. 2009. PMID: 19019143 Free PMC article.
-
Maintenance of chromosome structure in Pseudomonas aeruginosa.FEMS Microbiol Lett. 2014 Jul;356(2):154-65. doi: 10.1111/1574-6968.12478. Epub 2014 Jun 12. FEMS Microbiol Lett. 2014. PMID: 24863732 Free PMC article. Review.
-
Regulating DnaA complex assembly: it is time to fill the gaps.Curr Opin Microbiol. 2010 Dec;13(6):766-72. doi: 10.1016/j.mib.2010.10.001. Epub 2010 Oct 27. Curr Opin Microbiol. 2010. PMID: 21035377 Free PMC article. Review.
References
-
- Atlung, T., E. Clausen, and F. G. Hansen. 1985. Autoregulation of the dnaA gene of Escherichia coli. Mol. Gen. Genet. 200:442-450. - PubMed
-
- Atlung, T., L. J. Rasmussen, L. J. Nellemann, and F. Holm. 1991. A versatile method for integration of genes and gene fusions into the L(lambda) attachment site of Escherichia coli. Gene 107:11-17. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

