The Escherichia coli CpxA-CpxR envelope stress response system regulates expression of the porins ompF and ompC
- PMID: 16077119
- PMCID: PMC1196077
- DOI: 10.1128/JB.187.16.5723-5731.2005
The Escherichia coli CpxA-CpxR envelope stress response system regulates expression of the porins ompF and ompC
Abstract
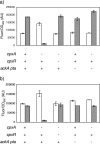
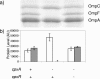
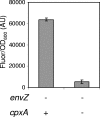
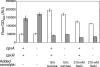
We performed transposon mutagenesis of a two-color fluorescent reporter strain to identify new regulators of the porin genes ompF and ompC in Escherichia coli. Screening of colonies by fluorescence microscopy revealed numerous mutants that exhibited interesting patterns of porin expression. One mutant harbored an insertion in the gene encoding the histidine kinase CpxA, the sensor for a two-component signaling system that responds to envelope stress. The cpxA mutant exhibited increased transcription of ompC and a very strong decrease in transcription of ompF under conditions in which acetyl phosphate levels were high. Subsequent genetic analysis revealed that this phenotype is dependent on phosphorylation of the response regulator CpxR and that activation of CpxA in wild-type cells results in similar regulation of porin expression. Using DNase I footprinting, we demonstrated that CpxR binds upstream of both the ompF and ompC promoters. It thus appears that two distinct two-component systems, CpxA-CpxR and EnvZ-OmpR, converge at the porin promoters. Within the context of envelope stress, outer membrane beta-barrel proteins have generally been associated with the sigma E pathway. However, at least for the classical porins OmpF and OmpC, our results show that the Cpx envelope stress response system plays a role in regulating their expression.
Figures






References
-
- Ades, S. E. 2004. Control of the alternative sigma factor sigmaE in Escherichia coli. Curr. Opin. Microbiol. 7:157-162. - PubMed
-
- Alba, B. M., and C. A. Gross. 2004. Regulation of the Escherichia coli sigma-dependent envelope stress response. Mol. Microbiol. 52:613-619. - PubMed
-
- Amann, E., B. Ochs, and K. J. Abel. 1988. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 69:301-315. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

