Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins
- PMID: 16079181
- PMCID: PMC1237069
- DOI: 10.1091/mbc.e05-01-0033
Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins
Abstract
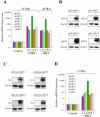
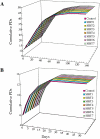
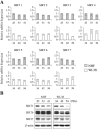
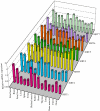
Sir2 is a NAD+-dependent protein deacetylase that extends lifespan in yeast and worms. This study examines seven human proteins homologous to Sir2 (SIRT1 through SIRT7) for cellular localization, expression profiles, protein deacetylation activity, and effects on human cell lifespan. We found that: 1) three nuclear SIRT proteins (SIRT1, SIRT6, and SIRT7) show different subnuclear localizations: SIRT6 and SIRT7 are associated with heterochromatic regions and nucleoli, respectively, where yeast Sir2 functions; 2) SIRT3, SIRT4, and SIRT5 are localized in mitochondria, an organelle that links aging and energy metabolism; 3) cellular p53 is a major in vivo substrate of SIRT1 deacetylase, but not the other six SIRT proteins; 4) SIRT1, but not the other two nuclear SIRT proteins, shows an in vitro deacetylase activity on histone H4 and p53 peptides; and 5) overexpression of any one of the seven SIRT proteins does not extend cellular replicative lifespan in normal human fibroblasts or prostate epithelial cells. This study supports the notion that multiple human SIRT proteins have evolutionarily conserved and nonconserved functions at different cellular locations and reveals that the lifespan of normal human cells, in contrast to that of lower eukaryotes, cannot be manipulated by increased expression of a single SIRT protein.
Figures




References
-
- Baptiste, N., and Prives, C. (2004). p53 in the cytoplasm: a question of overkill? Cell 116, 487–489. - PubMed
-
- Barbieri, M., Bonafe, M., Franceschi, C., and Paolisso, G. (2003). Insulin/IGF-I-signaling pathway: an evolutionarily conserved mechanism of longevity from yeast to humans. Am. J. Physiol. Endocrinol. Metab. 285, E1064–E1071. - PubMed
-
- Blander, G., and Guarente, L. (2004). The sir2 family of protein deacetylases. Annu. Rev. Biochem. 73, 417–435. - PubMed
-
- Bodnar, A. G., Ouellette, M., Frolkis, M., Holt, S. E., Chiu, C. P., Morin, G. B., Harley, C. B., Shay, J. W., Lichtsteiner, S., and Wright, W. E. (1998). Extension of life-span by introduction of telomerase into normal human cells. Science 279, 349–352. - PubMed
-
- Brooks, C. L., and Gu, W. (2003). Ubiquitination, phosphorylation and acetylation: the molecular basis for p53 regulation. Curr. Opin. Cell Biol. 15, 164–171. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

