Genetic and genomic analysis of the AT-rich centromere DNA element II of Saccharomyces cerevisiae
- PMID: 16079225
- PMCID: PMC1350974
- DOI: 10.1534/genetics.105.046458
Genetic and genomic analysis of the AT-rich centromere DNA element II of Saccharomyces cerevisiae
Abstract
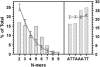
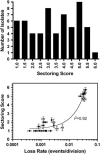
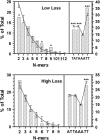
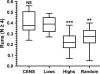
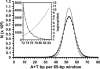
Centromere DNA element II (CDEII) of budding yeast centromeres is an AT-rich sequence essential for centromere (CEN) function. Sequence analysis of Saccharomyces cerevisiae CDEIIs revealed that A(5-7)/T(5-7) tracts are statistically overrepresented at the expense of AA/TT and alternating AT. To test the hypothesis that this nonrandom sequence organization is functionally important, a CEN library in which the CDEII sequences were randomized was generated. The library was screened for functional and nonfunctional members following centromere replacement in vivo. Functional CENs contained CDEIIs with the highly biased A(n)/T(n) run distribution of native centromeres, while nonfunctional CDEIIs resembled those picked from the library at random. Run content, defined as the fraction of residues present in runs of four or more nucleotides, of the functional and nonfunctional CDEII populations differed significantly (P < 0.001). Computer searches of the genome for regions with an A + T content comparable to CDEIIs revealed that such loci are not unique to centromeres, but for 14 of the 16 chromosomes the AT-rich locus with the highest A(n > or =4) + T(n > or =4) run content was the centromere. Thus, the distinctive and nonrandom sequence organization of CDEII is important for centromere function and possesses informational content that could contribute to the determination of centromere identity.
Figures


References
-
- Brown, M. T., 1995. Sequence similarities between the yeast chromosome segregation protein Mif2 and the mammalian centromere protein CENP-C. Gene 160: 111–116. - PubMed
-
- Carbon, J., and L. Clarke, 1984. Structural and functional analysis of a yeast centromere (CEN3). J. Cell Sci. Suppl. 1: 43–58. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

