Interactions among DNA ligase I, the flap endonuclease and proliferating cell nuclear antigen in the expansion and contraction of CAG repeat tracts in yeast
- PMID: 16079237
- PMCID: PMC1456850
- DOI: 10.1534/genetics.105.043448
Interactions among DNA ligase I, the flap endonuclease and proliferating cell nuclear antigen in the expansion and contraction of CAG repeat tracts in yeast
Abstract
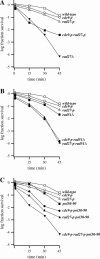
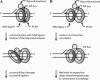
Among replication mutations that destabilize CAG repeat tracts, mutations of RAD27, encoding the flap endonuclease, and CDC9, encoding DNA ligase I, increase the incidence of repeat tract expansions to the greatest extent. Both enzymes bind to proliferating cell nuclear antigen (PCNA). To understand whether weakening their interactions leads to CAG repeat tract expansions, we have employed alleles named rad27-p and cdc9-p that have orthologous alterations in their respective PCNA interaction peptide (PIP) box. Also, we employed the PCNA allele pol30-90, which has changes within its hydrophobic pocket that interact with the PIP box. All three alleles destabilize a long CAG repeat tract and yield more tract contractions than expansions. Combining rad27-p with cdc9-p increases the expansion frequency above the sum of the numbers recorded in the individual mutants. A similar additive increase in tract expansions occurs in the rad27-p pol30-90 double mutant but not in the cdc9-p pol30-90 double mutant. The frequency of contractions rises in all three double mutants to nearly the same extent. These results suggest that PCNA mediates the entry of the flap endonuclease and DNA ligase I into the process of Okazaki fragment joining, and this ordered entry is necessary to prevent CAG repeat tract expansions.
Figures

References
-
- Bae, S.-H., E. Choi, K.-H. Lee, J. S. Park, S.-H. Lee et al., 1998. Dna2 of Saccharomyces cerevisiae possess a single-stranded DNA-specific endonuclease activity that is able to act on double-stranded DNA in the presence of ATP. J. Biol. Chem. 273: 26880–26890. - PubMed
-
- Bambara, R. A., R. S. Murante and L. A. Henricksen, 1997. Enzymes and reactions at the eukaryotic replication fork. J. Biol. Chem. 272: 4647–4650. - PubMed
-
- Bielinsky, A. K., and S. A. Gerbi, 1999. Chromosomal ARS1 has a single leading strand start site. Mol. Cell 3: 477–486. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

