Identifying gene-independent noncoding functional elements in the yeast ribosomal DNA by phylogenetic footprinting
- PMID: 16081534
- PMCID: PMC1182552
- DOI: 10.1073/pnas.0504905102
Identifying gene-independent noncoding functional elements in the yeast ribosomal DNA by phylogenetic footprinting
Abstract
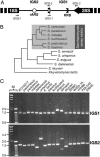
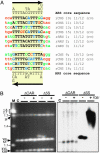
Sequences involved in the regulation of genetic and genomic processes are primarily located in noncoding regions. Identifying such cis-acting sequences from sequence data is difficult because their patterns are not readily apparent, and, to date, identification has concentrated on regions associated with gene-coding functions. Here, we used phylogenetic footprinting to look for gene-independent noncoding elements in the ribosomal RNA gene repeats from several Saccharomyces species. Similarity plots of ribosomal intergenic spacer alignments from six closely related Saccharomyces species allowed the identification of previously characterized functional elements, such as the origin-of-replication and replication-fork barrier sites, demonstrating that this method is a powerful predictor of noncoding functional elements. Seventeen previously uncharacterized elements, showing high levels of conservation, were also discovered. The conservation of these elements suggests that they are functional, and we demonstrate the functionality of two classes of these elements, a putative bidirectional noncoding promoter and a series of conserved peaks with matches to the origin-of-replication core consensus. Our results paint a comprehensive picture of the functionality of the Saccharomyces ribosomal intergenic region and demonstrate that functional elements not involved in gene-coding function can be identified by using comparative genomics based on sequence conservation.
Figures

References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

