Simultaneous visual detection of multiple viral amplicons by dipstick assay
- PMID: 16081944
- PMCID: PMC1233981
- DOI: 10.1128/JCM.43.8.4015-4021.2005
Simultaneous visual detection of multiple viral amplicons by dipstick assay
Abstract
A sensitive, simple, and instrument-independent method for the visual detection and identification of multiple nucleic acid amplicons by dipstick has been developed. This method is based on nucleic acid hybridization on the dipstick membrane and a signal amplification system to allow visual detection. With hepatitis B virus (HBV), hepatitis C virus (HCV), and human immunodeficiency virus type 1 (HIV-1) as model analytes, it is demonstrated that the visual dipstick test combined with multiplex reverse transcription (RT)-PCR for the amplification of viral nucleic acid provides a specific and sensitive detection method. The RT-PCR products were detected by the dipstick with an efficiency similar to that of a complex, expensive, and instrument-dependent method based on fluorogenic oligonucleotide probes. The detection limits of the dipstick combined with multiplex RT-PCR were 50, 125, and 500 IU/ml for HBV DNA, HCV RNA, and HIV-1 RNA, respectively. The dipstick assay detected with similar efficiencies amplicons derived from strains of HBV genotypes A through F, HCV genotypes 1 to 6, and HIV-1 subtypes A through H as well as CRF02 circulating recombinant forms of HIV-1. Analysis of 295 clinical samples and 19 pools of 10 plasma specimens from blood donors revealed that multiplex dipstick detection was reproducible, sensitive, and specific. The visual dipstick detection of multiple amplicons thus provides an attractive alternative to complex, instrument-dependent detection methods currently in use for nucleic acid testing. This new and sensitive method for nucleic acid detection should increase the availability of genomic screening in resource-limited settings and its applicability to near-patient testing.
Figures
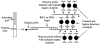
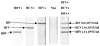

Comment in
-
Visual detection of multiple viral amplicons by dipstick assay: its application in screening of blood donors a welcome tool for the limited resource settings.J Clin Microbiol. 2005 Dec;43(12):6218; author reply 6218-9. doi: 10.1128/JCM.43.12.6218-6219.2005. J Clin Microbiol. 2005. PMID: 16333138 Free PMC article. No abstract available.
References
-
- Allain, J.-P., P. E. Hewitt, R. S. Tedder, and L. M. Williamson. 1999. Evidence that anti-HBc but not HBV DNA testing may prevent some HBV transmission by transfusion. Br. J. Haematol. 107:186-195. - PubMed
-
- Allain, J.-P., D. Candotti, K. Soldan, F. Sarkodie, B. Phelps, C. Giachetti, V. Shyamala, F. Yeboah, M. Anokwa, S. Owusu-Ofori, and O. Opare-Sem. 2003. The risk of hepatitis B virus infection by transfusion in Kumasi, Ghana. Blood 101:2419-2425. - PubMed
-
- Allain, J.-P. 2004. Occult hepatitis B virus infection: implications in transfusion. Vox Sang. 86:83-91. - PubMed
-
- Baeumner, A. J., N. A. Schlesinger, N. S. Slutzki, J. Romano, E. M. Lee, and R. A. Montagna. 2002. Biosensor for dengue virus detection: sensitive, rapid, and serotype specific. Anal. Chem. 74:1442-1448. - PubMed
-
- Biswas, R., E. Tabor, C. C. Hsia, D. J. Wright, M. E. Laycock, E. W. Fiebig, L. Peddada, R. Smith, G. B. Schreiber, J. S. Epstein, G. J. Nemo, and M. P. Busch. 2003. Comparative sensitivity of HBV NATs and HBsAg assays for detection of acute HBV infection. Transfusion 43:788-798. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

