Distribution of the serine protease autotransporters of the Enterobacteriaceae among extraintestinal clinical isolates of Escherichia coli
- PMID: 16081954
- PMCID: PMC1233896
- DOI: 10.1128/JCM.43.8.4076-4082.2005
Distribution of the serine protease autotransporters of the Enterobacteriaceae among extraintestinal clinical isolates of Escherichia coli
Abstract
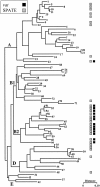
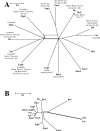
Urinary tract infections continue to be among the most common extraintestinal diseases. Cystitis in women is by far the most common urinary tract infection; pyelonephritis in both sexes and prostatitis in men are more severe but less frequent complaints. Escherichia coli is by far the most common cause of urinary tract infection. It is believed that uropathogenic E. coli is adept at colonizing the urinary tract via the production of specific virulence factors. Recently, a novel virulence determinant, Vat, was described for the prototypical uropathogenic E. coli strain CFT073. Vat is a member of the SPATE (serine protease autotransporters of the Enterobacteriaceae) subfamily of the autotransporters. Previously, SPATEs have been described for all pathovars of E. coli, but until recently their presence had been noticeably absent in nonpathogenic E. coli. In this report we describe the prevalence and phylogenetic distribution of the SPATEs among uropathogenic E. coli and the ECOR collection, demonstrating an association between the presence of the SPATEs, including Vat, and uropathogenic E. coli phylogroups. In addition, we describe the distribution of SPATEs among nonpathogenic E. coli.
Figures

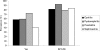
Similar articles
-
The high prevalence of serine protease autotransporters of Enterobacteriaceae (SPATEs) in Escherichia coli causing neonatal septicemia.Eur J Clin Microbiol Infect Dis. 2014 Nov;33(11):2015-24. doi: 10.1007/s10096-014-2161-4. Epub 2014 Jun 13. Eur J Clin Microbiol Infect Dis. 2014. PMID: 24924922
-
Three new serine-protease autotransporters of Enterobacteriaceae (SPATEs) from extra-intestinal pathogenic Escherichia coli and combined role of SPATEs for cytotoxicity and colonization of the mouse kidney.Virulence. 2019 Dec;10(1):568-587. doi: 10.1080/21505594.2019.1624102. Virulence. 2019. PMID: 31198092 Free PMC article.
-
Distribution of serine protease autotransporters of Enterobacteriaceae in typical and atypical enteroaggregative Escherichia coli.Infect Genet Evol. 2017 Jun;50:83-86. doi: 10.1016/j.meegid.2017.02.018. Epub 2017 Feb 28. Infect Genet Evol. 2017. PMID: 28254427
-
Serine Protease Autotransporters of the Enterobacteriaceae (SPATEs): Out and About and Chopping It Up.Microorganisms. 2019 Nov 21;7(12):594. doi: 10.3390/microorganisms7120594. Microorganisms. 2019. PMID: 31766493 Free PMC article. Review.
-
Serine protease autotransporters of enterobacteriaceae (SPATEs): biogenesis and function.Toxins (Basel). 2010 Jun;2(6):1179-206. doi: 10.3390/toxins2061179. Epub 2010 May 28. Toxins (Basel). 2010. PMID: 22069633 Free PMC article. Review.
Cited by
-
Increasing Fluroquinolone Susceptibility and Genetic Diversity of ESBL-Producing E. coli from the Lower Respiratory Tract during the COVID-19 Pandemic.Antibiotics (Basel). 2024 Aug 23;13(9):797. doi: 10.3390/antibiotics13090797. Antibiotics (Basel). 2024. PMID: 39334972 Free PMC article.
-
Pathogenicity Factors of Genomic Islands in Intestinal and Extraintestinal Escherichia coli.Front Microbiol. 2020 Sep 25;11:2065. doi: 10.3389/fmicb.2020.02065. eCollection 2020. Front Microbiol. 2020. PMID: 33101219 Free PMC article. Review.
-
Molecular Characterization of the Vacuolating Autotransporter Toxin in Uropathogenic Escherichia coli.J Bacteriol. 2016 Apr 28;198(10):1487-98. doi: 10.1128/JB.00791-15. Print 2016 May 15. J Bacteriol. 2016. PMID: 26858103 Free PMC article.
-
Prevalence and changing antimicrobial resistance profiles of Shigella spp. isolated from diarrheal patients in Kolkata during 2011-2019.PLoS Negl Trop Dis. 2024 Feb 20;18(2):e0011964. doi: 10.1371/journal.pntd.0011964. eCollection 2024 Feb. PLoS Negl Trop Dis. 2024. PMID: 38377151 Free PMC article.
-
Genomic Insights into Drug Resistance and Virulence Platforms, CRISPR-Cas Systems and Phylogeny of Commensal E. coli from Wildlife.Microorganisms. 2021 May 5;9(5):999. doi: 10.3390/microorganisms9050999. Microorganisms. 2021. PMID: 34063152 Free PMC article.
References
-
- Bauer, R. J., L. Zhang, B. Foxman, A. Siitonen, M. E. Jantunen, H. Saxen, and C. F. Marrs. 2002. Molecular epidemiology of 3 putative virulence genes for Escherichia coli urinary tract infection-usp, iha, and iron (E. coli). J. Infect. Dis. 185:1521-1524. - PubMed
-
- Benjelloun-Touimi, Z., P. J. Sansonetti, and C. Parsot. 1995. SepA, the major extracellular protein of Shigella flexneri: autonomous secretion and involvement in tissue invasion. Mol. Microbiol. 17:123-135. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

